Research Articles:
Microbial Cell, Vol. 4, No. 9, pp. 294 - 304; doi: 10.15698/mic2017.09.589
Farnesol inhibits translation to limit growth and filamentation in C. albicans and S. cerevisiae
1 Division of Molecular and Cellular Function, School of Biological Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester Academic Health Science Centre, Michael Smith Building, Oxford Rd., Manchester, M13 9PT, United Kingdom.
2 Current address: Department of Biological Sciences, Nigerian Defence Academy, PMB 2109, Kaduna, Nigeria.
3 Current address: Sheffield Institute for Translational Neuroscience, University of Sheffield, Sheffield, S10 2HQ, United Kingdom.
Keywords: protein synthesis, translation control, farnesol, quorum sensing.
Received originally: 21/04/2017 Received in revised form: 01/08/2017
Accepted: 13/08/2017
Published: 04/09/2017
Correspondence:
Mark P. Ashe, Division of Molecular and Cellular Function, School of Biological Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester Academic Health Science Centre, Michael Smith Building, Oxford Rd., Manchester, M13 9PT, United Kingdom; Tel: +44 (0)161 306 4164; mark.p.ashe@manchester.ac.uk
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Nkechi E. Egbe, Tawni O. Dornelles, Caroline M. Paget, Lydia M. Castelli and Mark P. Ashe (2017). Farnesol inhibits translation to limit growth and filamentation in C. albicans and S. cerevisiae. Microbial Cell 4(9): 294-304. doi: 10.15698/mic2017.09.589
Abstract
Candida albicans is a polymorphic yeast where the capacity to switch between yeast and filamentous growth is critical for pathogenicity. Farnesol is a quorum-sensing sesquiterpene alcohol that, via regulation of specific signalling and transcription components, inhibits filamentous growth in C. albicans. Here we show that farnesol also inhibits translation at the initiation step in both C. albicans and S. cerevisiae. In contrast to fusel alcohols, that target the eukaryotic initiation factor 2B (eIF2B), farnesol affects the interaction of the mRNA with the small ribosomal subunit leading to reduced levels of the 48S preinitiation ribosomal complex in S. cerevisiae. Therefore, farnesol targets a different step in the translation pathway than fusel alcohols to elicit a completely opposite physiological outcome by negating filamentous growth.
INTRODUCTION
The capacity to detect and respond to environmental change is essential for microorganism survival. This is especially true for opportunist pathogens like Candida albicans; where to initiate infection, the organism must adapt and persist in spite of host immune responses. Typically, C. albicans is a harmless commensal, yet in infected patients it causes various different conditions, from mucosal infections to life-threatening systemic infections [1].
–
Cell-cell signalling, particularly quorum sensing (QS), is a major focus of microbiological research. Farnesol is an acyclic sesquiterpene alcohol that represents the first QS molecule identified in eukaryotic microorganisms [2] where it causes a range of physiological effects [3]. In C. albicans, farnesol inhibits the yeast to hyphal switch [2] to prevent colonization of different niche environments [4][5], it has antioxidant effects [6][7] and it inhibits transporters [8]. In many species farnesol induces cellular death: for example in the fungal species, Saccharomyces cerevisiae [9], Aspergillus nidulans [10], Penicillium expansum [11], Botrytis cinerea [12] and even C. albicans under certain conditions [13]. Equally, farnesol triggers cell death in mammalian cells [14] and can have antibacterial properties [15][16]. In fact, farnesol was first discovered as a constituent of plant essential oils with antimicrobial activities [17].
–
Cellular responses to stimuli act via signal transduction pathways to regulate gene expression. In C. albicans, farnesol targets pathways like the Ras-PKA pathway that, via the transcription factors Efg1p and Czf1p and the repressor Tup1p, regulates gene expression [18]. If a stimulus induces cellular stress, a transient inhibition of global protein synthesis is often observed, which further modulates the programme of gene expression to allow stress responsive gene expression programs to be initiated [19][20]. Control of translation in this manner mostly occurs at the initiation stage in order to allow rapid and reversible management of gene expression.
–
Translation initiation is the assembly of an elongation competent 80S ribosome with an initiator methionyl-tRNA (Met-tRNAiMet) base paired via its anticodon loop to an mRNA Start codon [21]. Highly conserved controls allow eukaryotic cells to globally reduce translation [19][20]: a prominent example involves eIF2α kinases, like Gcn2p in S. cerevisiae [22][23]. Gcn2p activation after amino acid starvation causes phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF2) [24]. eIF2 is an essential GTP-binding protein that interacts with Met-tRNAiMet to form a ternary complex (TC) that is competent for initiating translation [23]. Phosphorylated eIF2 competitively inhibits the eIF2B-mediated guanine nucleotide exchange reaction on eIF2, reducing TC levels and translation initiation [23]. However, specific mRNAs, such as yeast GCN4, continue to be translated under these conditions. GCN4 encodes a transcription factor that regulates the expression of amino acid biosynthetic genes. This feedback regulatory circuit has proved a paradigm for studies on translation control [25]. Similarly, C. albicans expresses a single eIF2α kinase, Gcn2p, which phosphorylates eIF2α in response to various stresses [26][27] and translational activation of CaGCN4 also provides feedback regulation [28][29].
–
As well as indirect attenuation of eIF2B activity via phosphorylation of eIF2α, cells can also modulate eIF2B activity more directly. In mammalian cells, phosphorylation of eIF2B has been identified as an important regulatory mechanism [30]. In addition, in both yeast and mammalian cells volatile anaesthetics appear to inhibit protein synthesis via eIF2B regulation [31][32]. Moreover, in both S. cerevisiae and C. albicans, fusel alcohols, which are also characterised as quorum sensing molecules [33], have been shown to inhibit translation initiation in a mechanism that targets eIF2B but independently of the Gcn2p kinase or eIF2α phosphorylation [34][35][36].
–
Besides control via eIF2B, another regulated step in translation initiation is the mRNA selection phase [37]. eIF4E and Pab1p select mRNA via interaction with the 5ʹ cap and 3ʹ poly(A) tail, respectively. eIF4G can interact with both eIF4E and Pab1p to form a closed loop complex that, via interactions with eIF3, eIF5 and eIF1, can recruit the small ribosomal subunit to form a 48S preinitiation complex [21]. A variety of stress conditions have been shown to target these steps in the initiation pathway leading to transient reductions in translation to facilitate a switch to a new program of gene expression [19][20].
–
In this study, we show that as well as hampering various filamentation pathways, farnesol inhibits protein synthesis. This inhibition of translation occurs at the initiation step and most likely impacts upon the assembly of the 48S preinitiation complex. Intriguingly, this means two different quorum sensing agents, farnesol and fusel alcohols that have conflicting effects on filamentous growth, both inhibit translation initiation but by different mechanisms.
RESULTS AND DISCUSSION
Farnesol inhibits growth and protein synthesis
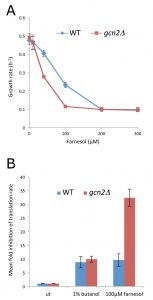
Farnesol, a eukaryotic QS molecule inhibits filamentous growth in both S. cerevisiae and C. albicans [2], however, the concentration required varies according to the specific growth regime [38]. Under the growth conditions used here, we found concentrations in excess of 100 µM farnesol inhibited the growth of C. albicans and an isogenic gcn2∆ mutant (Fig. 1A). To study possible origins of the growth inhibition, the impact of farnesol on the rate of protein synthesis was monitored. The resulting [35S]-methionine incorporation data show that farnesol (300 µM) and butanol (2%) cause a 10-fold inhibition of protein synthesis in the CAI4 strain of C. albicans (Fig. 1B). Therefore, farnesol inhibits protein synthesis at very early stages after addition and this control could contribute to the growth inhibition observed.
–
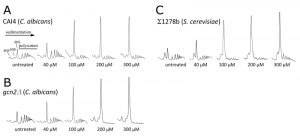
To further investigate the stage of protein synthesis that is targeted by farnesol, polysome profiling was used, as this allows both the level of protein synthesis and the stage of regulation to be investigated [34][35][36]. Analysis of polysome distribution for the C. albicans CAI4 strain revealed that increasing concentrations of farnesol caused a change in the polysome profile (Fig. 2A). The 80S peak increased dramatically and the polysome peaks were reduced. This change in profile is characteristic of an inhibition of translation initiation [34] and has been observed for many stresses [19]. Similar results in terms of farnesol sensitivity were obtained for the Σ1278b strain of S. cerevisiae, where similar concentrations elicited the response across the two yeast species (Fig. 2C). It has been noted previously that the level of free 60S is particularly high for the Σ1278b, although this does not appear to impact upon its growth or its sensitivity to translational stress [39]. The lowest farnesol concentration that caused a gross impact on polysome distribution for either C. albicans or S. cerevisiae was 100 mM (Fig. 2). This correlated well with the concentration that inhibited growth under the conditions used here (Fig. 1A) suggesting that the inhibition of translation initiation could be intrinsically connected to growth inhibition for farnesol.
–
The CAI4 strain of C. albicans used here is a commonly used lab strain that is auxotrophic in the uracil biosynthetic pathway by virtue of a homozygous deletion of the URA3 gene [40]. This mutation has previously been shown to alter a number of aspects of C. albicans physiology including adhesion and virulence [41]. Therefore, a prototrophic strain of C. albicans, SC5314, was tested. Entirely analogous observations were made on the impact of farnesol on the growth (Fig. S1A) and translation (Fig. S1B) of this strain in response to similar concentrations of farnesol.
–
Previous studies evaluating QS have established that trans,trans-farnesol is produced by Candida as a QS molecule to inhibit filamentation [2][42]. Intriguingly, the trans,trans form and mixed stereoisomer preparations both impact upon translation initiation equally (Fig. S1). Indeed 40 mM of each is sufficient to induce a mild inhibition of protein synthesis and 100 mM leads to a robust inhibition (Fig. 1B). Therefore, in order to explore the mechanism by which farnesol inhibits translation initiation, over the course of the rest of our studies 100 mM farnesol was used as this concentration elicits robust inhibition of both growth and translation. However, it should be noted that lower concentrations of farnesol (e.g. 40 mM) can lead to subtle alterations in the polysome profile (Fig. 2, 3 and S1). This level of sensitivity to farnesol correlates well with earlier studies using similar growth conditions [38].
–
GCN2 is not involved in the inhibition translation initiation by farnesol
In terms of the mechanism of translational regulation, like S. cerevisiae, C. albicans harbours a single eIF2a kinase gene, GCN2, which is involved in the regulation of translation initiation in response to various stresses [26][27]. Indeed in S. cerevisiae, gcn2Δ mutants are incapable of inhibiting translation initiation in response to specific stress conditions [34], which prevents cells from mounting an appropriate stress response. Therefore, the role of Gcn2p in the farnesol-dependent inhibition of translation initiation was assessed using a gcn2Δ mutant strain of C. albicans [26]. Previous observations using this strain show that translation initiation remains uninhibited early after amino acid starvation [29]. In terms of the impact of farnesol on growth and translation, the gcn2∆ mutant strain is at least as sensitive as the wild type (Fig. 1A and B). In fact rather than the gcn2∆ mutant being resistant to farnesol in terms of translation, as might be expected if the Gcn2p kinase were involved in the control, the farnesol-treated gcn2∆ strain is even more inhibited than the wild type. Growth is inhibited at lower farnesol concentrations for the gcn2∆ mutant and methionine incorporation is inhibited up to 30-fold (Fig. 1A and B). Equally, a comparison of the polysome profiles shows that the gcn2∆ mutant exhibits somewhat greater sensitivity than the wild type mirroring the growth phenotypes (Fig. 2B). For instance, following treatment with 300 mM farnesol, greater polysome run-off is observed for the gcn2∆ mutant compared to the parent strain (cf. Fig. 2B with 2A). Overall, these results show that the Gcn2p kinase, that is a requirement for eIF2a phosphorylation and the subsequent regulation of translation initiation in response to a variety of stresses, is not required for the inhibition of translation initiation by farnesol in C. albicans; in fact the gcn2∆ mutant is more sensitive to treatment.
–
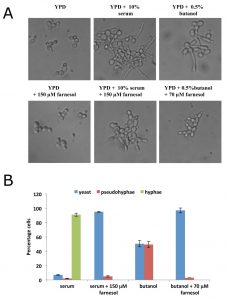
Fusel alcohols and other conditions that inhibit protein synthesis in C. albicans promote filamentous growth [36], whereas farnesol inhibits protein synthesis and prevents filamentation. An obvious query is whether the filamentation inducing signal generated by fusel alcohols can be overridden by farnesol or vice versa. Induction of filamentous growth is further complicated as different cues induce distinct forms of filamentation [43]. For instance, fusel alcohols are characterised as inducing pseudohyphal growth [44] where elongated, ellipsoid yeast cells remain attached to one another via constricted septation sites leading to growth of a colony in a branched pattern [45]. In contrast, serum addition elicits true hyphal growth [46], whereby cells are narrow, long, have parallel sides and no obvious constrictions points [45]. In the presence of serum alone over 90% hyphal growth was observed and the addition of 150 µM farnesol blocked the yeast to hyphae switch (Fig. 3A and B). In contrast, the fusel alcohol, butanol, induces a much less robust effect whereby roughly 50% of cells exhibit pseudohyphal morphology. Here just 70 µM farnesol was sufficient to block any filamentous growth. These results show that farnesol competes with both serum and butanol, but the concentration of farnesol required to effect competition varies according to the strength of the filamentation signal (Fig. 3A and B). Curiously, even though both fusel alcohols and farnesol target protein synthesis, they are in competition with respect to their physiological impact on filamentous growth. Thus previous observations suggesting that the inhibition of protein synthesis favours filamentation [36] cannot be generalised across all conditions.
–
Eukaryotic initiation factor 2B (eIF2B) is not regulated by farnesol
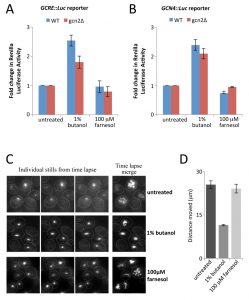
Fusel alcohols inhibit translation initiation in S. cerevisiae and C. albicans by targeting the guanine nucleotide exchange factor, eIF2B leading to reduced levels of the eIF2•GTP•Met-tRNAi ternary complex [34][35][36]. Specific general control response reporters such as the GCN4 reporter mRNA provide a sensitive indicator of changes in the ternary complex and are widely used to study translational regulation [25]. A key observation that pointed towards eIF2B as a target for fusel alcohols was the demonstration that these reporters of ternary complex levels are translationally up-regulated [34][36]. In order to assess this response after farnesol treatment, strains carrying two renilla luciferase reporters were used: the first contains five copies of the general control response element (GCRE), while the second harbours the GCN4 promoter and leader region upstream [28][29]. Using the parent and gcn2∆ mutant strains bearing these reporters, the previous observation that 1% butanol elicits a non-Gcn2p dependent increase in the activity of GCRE-Luc and GCN4-Luc was confirmed (Fig. 4A and B). In stark contrast, farnesol elicits no significant increases of the GCRE-Luc or GCN4-Luc reporter expression (Fig. 4A and B) suggesting that farnesol does not alter ternary complex levels to activate the GCN response.
–
Previous studies on the localisation of eIF2B in organisms from S. cerevisiae to Drosophila melanogaster have defined a large cytoplasmic body called the eIF2B body (2B body) [35][36][47][48][49]. Exposure of either S. cerevisiae or C. albicans cells to fusel alcohols reduces the dynamics of this 2B body in a manner that correlates with the sensitivity/ resistance of strains to alcohols [35][36]. In order to ascertain whether farnesol also impacts upon 2B body dynamics, a C. albicans strain bearing GFP-tagged eIF2Bγ was used [36]. Epiflourescence time-lapse microscopy experiments were performed by acquiring images of untreated, butanol treated or farnesol treated cells over a 2 min period. Movement of the eIF2B body across the images was tracked and the total distance (µm) moved was calculated. Quantitation of the average displacement shows that 1% butanol causes total eIF2B body movement to drop by approximately 50%. In contrast, in farnesol treated cells the 2B body moves to the same extent as in the untreated cells (Fig. 4C and D). This observation again suggests that the regulatory mechanism by which farnesol inhibits translation initiation is distinct from that of fusel alcohols and is not dependent upon eIF2B regulation or the alteration of ternary complex levels.
–
Farnesol inhibits translation initiation by targeting 48S preinitiation complex formation
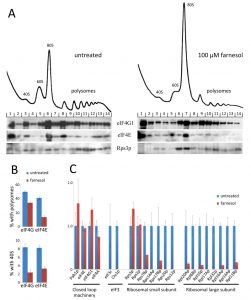
Translation initiation is controlled at other levels besides ternary complex formation and eIF2B. For instance, the interaction of mRNA with the 43S preinitiation complex, i.e. 48S preinitiation complex formation, can also be regulated [19][20][37]. This process relies upon interactions between proteins that bind the mRNA and proteins associated with the 40S ribosomal subunit complex. One way to assess the factors present with the 40S ribosomal subunit is to perform immunoblotting on fractions collected from across sucrose density polysomal gradients. Formaldehyde cross-linking prior to cell lysis stabilizes protein factors in such complexes during the subsequent sedimentation and fractionation steps [50][51].
–
A limitation of such studies in C. albicans is that many antibodies against translation factors that are available for S. cerevisiae do not cross-react with C. albicans proteins (data not shown). Therefore, to further investigate the step in the translation pathway that is targeted by farnesol, investigations were undertaken in S. cerevisiae. The Ʃ1278b laboratory strain was selected, as like C. albicans, Ʃ1278b is diploid and can undergo morphogenetic switching to pseudohyphal growth [44]. In order to validate the use of this strain, the effects of farnesol on growth and translation were cross-compared. Farnesol inhibits growth and translation initiation over a similar concentration range for the two yeasts and for other lab strains of S. cerevisiae, such as BY4741 (Fig. 2C; data not shown), so it seems likely that the translational responses to butanol and farnesol are mechanistically conserved across these species.
–
Formaldehyde polysome analysis after farnesol treatment revealed an interesting effect in terms of the region of the gradient harbouring the 40S ribosomal subunit. This region not only contains free 40S ribosomal subunits but also the 48S ribosomal preinitiation complex where the 40S subunit is associated with mRNA and translation initiation factors i.e. an intermediate in the translation initiation process. A marker for this complex is the presence of translation initiation factors such as eIF4E and eIF4G that are specifically targeted to the mRNA rather than the 40S ribosomal subunit. The level of both eIF4G1 and eIF4E in the 40S region decreased dramatically after farnesol treatment (Fig. 5A, cf. fraction 3 untreated and fraction 3 treated). Quantitation confirmed that levels dropped from ~8-9% of total to 2-3% after farnesol treatment (Fig. 5B). Furthermore, both eIF4G and eIF4E are reduced in polysome regions and this likely reflects reduced levels of initiating ribosomes on mRNAs that are already being translated: although it should be noted that the scale of reduction is greater for eIF4E than eIF4G (Fig. 5B). This may relate to the fact that eIF4G can interact with RNA, Pab1p and other translation factors, whereas eIF4E is targeted to the mRNA cap. In sum, these data highlight the possibility that farnesol causes an alteration in protein-protein interactions that lie upstream of 48S complex formation.
–
In order to further investigate how farnesol treatment leads to eIF4G/eIF4E depletion from regions of the gradient, we undertook an immunopurification-mass spectrometry strategy using an eIF4G1-TAP tagged S. cerevisiae strain. In terms of the relative number of peptides observed in the immunopurified samples, peptides from known mRNA associated factors, such as eIF4G, eIF4E, eIF4A and Pab1p, were largely unaffected by farnesol (Fig. 5B). In contrast, peptides for other components of the translation machinery were reduced dramatically; including peptides for the ribosomal proteins, as well as subunits of eIF3, a translation initiation factor that is associated with the 40S ribosomal subunit. Overall, these data support a model where farnesol targets the formation of the 48S preinitiation complex to inhibit protein synthesis. The fact that farnesol targets a different step in translation to fusel alcohols, may mean that the impact of these agents and the stage targeted contributes to the opposite effects in terms of filamentous growth (Fig. 3). This is suggestive that translational control plays an important role in the physiological response of C. albicans to QS molecules.
DISCUSSION
A range of alcohols or their derivatives can act as signalling molecules across yeast species [2][44][52][53]. The data presented here combined with that in our previous studies [36] show that both butanol and farnesol inhibit protein synthesis at the translation initiation stage in C. albicans. Both are metabolites of C. albicans that act as signalling molecules, yet have opposing effects on morphological transition [2][36]. Previously, we have shown in both S. cerevisiae and C. albicans that short chain alcohols regulate protein synthesis by targeting the guanine nucleotide exchange factor eIF2B [34][35][36].
–
eIF2B regulation plays a critical role in reprograming gene expression as part of the response to stress across different eukaryotic cells [23]. For instance, eIF2 phosphorylation by eIF2a kinases, like Gcn2p in yeast, inhibits eIF2B in response to stresses such as amino acid starvation [24], purine starvation [54] and rapamycin treatment [55]. However, while these stresses target eIF2B in a Gcn2p-dependent manner, the mechanism by which short chain alcohols target eIF2B in both S. cerevisiae and C. albicans is Gcn2p-independent [34][35][36]. The GCN4 reporter experiments and analysis of the eIF2B body in this study suggest that the longer chain sesquiterpene alcohol farnesol inhibits translation initiation in a mechanism that does not involve eIF2B regulation; either Gcn2p-dependent or independent.
–
Many studies have reported translational controls targeting steps upstream of 48S preinitiation complex formation. For instance, glucose starvation in S. cerevisiae causes a reorganisation of the closed loop mRNP translation complex, whereby eIF4A dissociates and the cosedimentation of eIF4E, eIF4G and Pab1p with ribosomal complexes is compromised [51]. The small non-coding BC RNAs in neuronal cells target the eIF4A helicase to inhibit 48S preinitiation complex formation on structured mRNAs [56]. Similarly, Burkholderia lethal factor 1, a toxin produced by Burkholderia pseudomallei, which causes the disease melioidosis, provokes a translational block via eIF4A [57]. 48S complex formation can also serve as the targeted step when specific mRNAs are translationally regulated. For instance, miRNAs have recently been shown to inhibit target mRNA translation by impacting upon eIF4A2 activity [58]. Therefore, a common translational regulatory mechanism that impacts upon the level of the 48S preinitiation complex is to target eIF4A activity.
–
In this study, we have investigated how farnesol effects different ribosomal complexes in S. cerevisiae using both formaldehyde-polysome analysis and immunoprecipitation followed by mass spectrometry. Both assays suggest that mRNA-associated translation factors (such as eIF4G, eIF4E and Pab1p) are associated less well with the ribosome and eIF3 following treatment with farnesol. Overall, the observed depletion of eIF4G and eIF4E from the 40S region of polysome gradients combined with the mass spectrometric analysis of eIF4G containing complexes lend support to a model where farnesol targets the formation of the 48S preinitiation complex to inhibit protein synthesis. This contrasts with the eIF2B dependent mechanism by which shorter chain alcohols target translation initiation.
–
Shorter chain alcohols and farnesol also differ in terms of their effects on morphological transitions in C. albicans. Short chain alcohols induce pseudohyphal growth in C. albicans whereas farnesol inhibits this process [2][44][53]. Indeed we show that farnesol can impede the filamentation induced by a variety of triggers including short-chain alcohols. This is not without precedent, in C. albicans farnesol also blocks morphogenesis induced by the aromatic alcohol, tyrosol [59]. One intriguing question is how both the shorter chain alcohols and farnesol can target a key ubiquitous process like protein synthesis, yet elicit distinct outcomes in terms of filamentous growth. This question drives at the fundamental physiological rationale for translational regulation in response to changing external conditions. Does the regulation constitute a knee-jerk reaction allowing the preservation of cellular resources by inhibiting the expression of the vast majority of mRNAs, or does the regulation serve a different purpose allowing specific mRNAs to be altered in their translation? In terms of farnesol, we show that translation is mildly inhibited at 40 mM farnesol and robustly down-regulated at 100 mM farnesol. Various Candida strains produce farnesol up to a concentration of ~60 mM [60], which would appear to favour the option where translation of a specific subset of mRNAs is altered. Evidence from a number of systems including the induction of GCN4 translation via amino acid starvation [25] would also favour this option. Under such a scenario, if two stresses impact upon different stages of translation initiation, they might alter the translation of different subsets of mRNA. We have previously observed evidence for such effects in S. cerevisiae, where fusel alcohols and amino acid starvation alter translational reprograming to allow continued translation of different cohorts of mRNAs [61]. With this in mind, we envisage that for farnesol and fusel alcohols, mRNAs encoding pro and anti-filamentation factors might be prominent in a set that are differentially regulated at the translational level. Such effects would also be integrated with well-defined transcriptional controls, especially for farnesol [18], to produce very different phenotypic outcomes.
MATERIALS AND METHODS
Media and growth conditions
The strains in Table 1 were grown and maintained as described previously [36]. Butanol and farnesol were routinely added for 15 min at the concentrations stated. Unless otherwise stated trans,trans-farnesol was used. Tolerance was assessed by adding butanol (0.5%, 1% and 2%) or farnesol (40 µM, 100 µM, 200 µM and 300 µM) to strains at OD600 0.1 and then testing growth.
 | TABLE 1. Strains used in this study. |
–
Morphogenesis assays
Exponential cultures were harvested, washed in water then re-inoculated into media with 0.5% butanol, 10% serum, 0.5% butanol – 70 µM farnesol or 10% serum – 150 µM farnesol. Filamentation was assessed microscopically as previously described [36].
–
Analysis of polysomes and other translation assays
Exponential strains were incubated with butanol/ farnesol for 15 min then treated with cycloheximide: 1 mg/ml (C. albicans) or 0.1 mg/ml (S. cerevisiae). Extracts were prepared then polysome analysis and fractionation were carried out as previous [36]. Formaldehyde polysome analysis was performed as described previously [50][51]. Immunoblots were probed with antibodies to yeast eIF4G, eIF4E and Rps3.
–
[35S]-methionine incorporation assays were conducted by adding 60 ng/ml methionine, where 0.5 ng/ml was [35S]-methionine (PerkinElmer), to exponential untreated or farnesol/ butanol treated cultures in synthetic complete dextrose (SCD) medium lacking methionine. Samples (1 ml) were taken at the indicated times and processed as described previously [36].
–
For the Luciferase reporter assays [29], lysates were prepared from exponential untreated or farnesol/ butanol treated cultures in RLUC buffer (0.5 M NaCl, 0.1 M K2HPO4, 1 mM Na2EDTA, 0.6 mM sodium azide, 1 mM phenylmethylsulfonyl fluoride, 0.02% bovine serum albumin). 1.25 μM coelentrazine h (Promega) was added to the extracts to initiate the reaction, and activity was measured using a GloMax 20/20 luminometer (Promega). Luciferase activity (RLU) is expressed as relative luminescence per 10 s/mg protein.
–
For studies on the 2B-body [35][36], real-time 2D deconvolved projections were generated via continuous z-sweep acquisition on a Delta Vision RT microscope (Applied Precision, Isaaquah, WA) with an Olympus 100× 1.40 NA DIC oil PlanApo objective (Melville, NY) and Roper CoolSnap HQ camera (Tucson, AZ) with Applied Precision Softworx 1.1 software for fast visualisation of all planes with minimal fluorescent bleaching. Images were acquired every 5 s over a 2 min period, and ImageJ (http://rsb.info.nih.gov/ij/; NIH) was used to track 2B body movement and calculate the mean total distance using at least 24 individual tracking experiments per condition.
–
Affinity Purification and mass spectrometry
For the eIF4G1-TAP purification, protein extracts were bound to IgG columns eluted with a TAP peptide, then samples were isolated from SDS PAGE gel slices [62]. Dried gel pieces containing the whole protein sample were digested using 100 ng trypsin and analysed by LC-MS/MS using an UltiMate® 3000 Rapid Separation liquid chromatography (Dionex Corporation, Sunnyvale, CA) coupled to a LTQ Velos Pro mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Data were searched using Mascot (Matrix Science UK), against the Uniprot database with S. cerevisiae selected. Data were validated and further processed using Scaffold (Proteome Software, Portland, OR).
References
- F.L. Mayer, D. Wilson, and B. Hube, "Candida albicanspathogenicity mechanisms", Virulence, vol. 4, pp. 119-128, 2013. http://dx.doi.org/10.4161/viru.22913
- J.M. Hornby, E.C. Jensen, A.D. Lisec, J.J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K.W. Nickerson, "Quorum Sensing in the Dimorphic Fungus Candida albicans Is Mediated by Farnesol", Applied and Environmental Microbiology, vol. 67, pp. 2982-2992, 2001. http://dx.doi.org/10.1128/AEM.67.7.2982-2992.2001
- M.L. Langford, A.L. Atkin, and K.W. Nickerson, "Cellular interactions of farnesol, a quorum-sensing molecule produced by Candida albicans", Future Microbiology, vol. 4, pp. 1353-1362, 2009. http://dx.doi.org/10.2217/fmb.09.98
- G. Ramage, S.P. Saville, B.L. Wickes, and J.L. López-Ribot, "Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule.", Applied and environmental microbiology, 2002. http://www.ncbi.nlm.nih.gov/pubmed/12406738
- M. Martins, M. Henriques, J. Azeredo, S.M. Rocha, M.A. Coimbra, and R. Oliveira, "Morphogenesis Control in Candida albicans and Candida dubliniensis through Signaling Molecules Produced by Planktonic and Biofilm Cells", Eukaryotic Cell, vol. 6, pp. 2429-2436, 2007. http://dx.doi.org/10.1128/EC.00252-07
- C. Westwater, E. Balish, and D.A. Schofield, "Candida albicans -Conditioned Medium Protects Yeast Cells from Oxidative Stress: a Possible Link between Quorum Sensing and Oxidative Stress Resistance", Eukaryotic Cell, vol. 4, pp. 1654-1661, 2005. http://dx.doi.org/10.1128/EC.4.10.1654-1661.2005
- A. Deveau, A.E. Piispanen, A.A. Jackson, and D.A. Hogan, "Farnesol Induces Hydrogen Peroxide Resistance in Candida albicans Yeast by Inhibiting the Ras-Cyclic AMP Signaling Pathway", Eukaryotic Cell, vol. 9, pp. 569-577, 2010. http://dx.doi.org/10.1128/EC.00321-09
- M. Sharma, and R. Prasad, "The Quorum-Sensing Molecule Farnesol Is a Modulator of Drug Efflux Mediated by ABC Multidrug Transporters and Synergizes with Drugs in Candida albicans", Antimicrobial Agents and Chemotherapy, vol. 55, pp. 4834-4843, 2011. http://dx.doi.org/10.1128/AAC.00344-11
- K. Machida, T. Tanaka, Y. Yano, S. Otani, and M. Taniguchi, "Farnesol-induced growth inhibition in Saccharomyces cerevisiae by a cell cycle mechanism", Microbiology, vol. 145, pp. 293-299, 1999. http://dx.doi.org/10.1099/13500872-145-2-293
- T. Dinamarco, M. Goldman, and G. Goldman, "Farnesol-induced cell death in the filamentous fungus Aspergillus nidulans", Biochemical Society Transactions, vol. 39, pp. 1544-1548, 2011. http://dx.doi.org/10.1042/BST0391544
- P. Liu, L. Luo, J. Guo, H. Liu, B. Wang, B. Deng, C. Long, and Y. Cheng, "Farnesol induces apoptosis and oxidative stress in the fungal pathogen Penicillium expansum.", Mycologia, 2010. http://www.ncbi.nlm.nih.gov/pubmed/20361499
- M. Cotoras, P. Castro, H. Vivanco, R. Melo, and L. Mendoza, "Farnesol induces apoptosis-like phenotype in the phytopathogenic fungusBotrytis cinerea", Mycologia, vol. 105, pp. 28-33, 2013. http://dx.doi.org/10.3852/12-012
- M.E. Shirtliff, B.P. Krom, R.A.M. Meijering, B.M. Peters, J. Zhu, M.A. Scheper, M.L. Harris, and M.A. Jabra-Rizk, "Farnesol-Induced Apoptosis in Candida albicans", Antimicrobial Agents and Chemotherapy, vol. 53, pp. 2392-2401, 2009. http://dx.doi.org/10.1128/AAC.01551-08
- J.H. Joo, and A.M. Jetten, "Molecular mechanisms involved in farnesol-induced apoptosis", Cancer Letters, vol. 287, pp. 123-135, 2010. http://dx.doi.org/10.1016/j.canlet.2009.05.015
- F.I.A. Gomes, P. Teixeira, J. Azeredo, and R. Oliveira, "Effect of Farnesol on Planktonic and Biofilm Cells of Staphylococcus epidermidis", Current Microbiology, vol. 59, pp. 118-122, 2009. http://dx.doi.org/10.1007/s00284-009-9408-9
- M.A. Jabra-Rizk, T.F. Meiller, C.E. James, and M.E. Shirtliff, "Effect of Farnesol on Staphylococcus aureus Biofilm Formation and Antimicrobial Susceptibility", Antimicrobial Agents and Chemotherapy, vol. 50, pp. 1463-1469, 2006. http://dx.doi.org/10.1128/AAC.50.4.1463-1469.2006
- V. Kuete, and T. Efferth, "Molecular determinants of cancer cell sensitivity and resistance towards the sesquiterpene farnesol.", Die Pharmazie, 2013. http://www.ncbi.nlm.nih.gov/pubmed/23923645
- M.L. Langford, J.C. Hargarten, K.D. Patefield, E. Marta, J.R. Blankenship, S. Fanning, K.W. Nickerson, and A.L. Atkin, "Candida albicans Czf1 and Efg1 Coordinate the Response to Farnesol during Quorum Sensing, White-Opaque Thermal Dimorphism, and Cell Death", Eukaryotic Cell, vol. 12, pp. 1281-1292, 2013. http://dx.doi.org/10.1128/EC.00311-12
- C. Simpson, and M. Ashe, "Adaptation to stress in yeast: to translate or not?", Biochemical Society Transactions, vol. 40, pp. 794-799, 2012. http://dx.doi.org/10.1042/BST20120078
- K.A. Spriggs, M. Bushell, and A.E. Willis, "Translational Regulation of Gene Expression during Conditions of Cell Stress", Molecular Cell, vol. 40, pp. 228-237, 2010. http://dx.doi.org/10.1016/j.molcel.2010.09.028
- A.G. Hinnebusch, "The Scanning Mechanism of Eukaryotic Translation Initiation", Annual Review of Biochemistry, vol. 83, pp. 779-812, 2014. http://dx.doi.org/10.1146/annurev-biochem-060713-035802
- G. Pavitt, "eIF2B, a mediator of general and gene-specific translational control", Biochemical Society Transactions, vol. 33, pp. 1487, 2005. http://dx.doi.org/10.1042/BST20051487
- R. Wek, H. Jiang, and T. Anthony, "Coping with stress: eIF2 kinases and translational control", Biochemical Society Transactions, vol. 34, pp. 7-11, 2006. http://dx.doi.org/10.1042/BST0340007
- T.E. Dever, L. Feng, R.C. Wek, A.M. Cigan, T.F. Donahue, and A.G. Hinnebusch, "Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast.", Cell, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1739968
- A.G. Hinnebusch, "TRANSLATIONAL REGULATION OFGCN4AND THE GENERAL AMINO ACID CONTROL OF YEAST", Annual Review of Microbiology, vol. 59, pp. 407-450, 2005. http://dx.doi.org/10.1146/annurev.micro.59.031805.133833
- H. Tournu, G. Tripathi, G. Bertram, S. Macaskill, A. Mavor, L. Walker, F.C. Odds, N.A.R. Gow, and A.J.P. Brown, "Global Role of the Protein Kinase Gcn2 in the Human Pathogen Candida albicans", Eukaryotic Cell, vol. 4, pp. 1687-1696, 2005. http://dx.doi.org/10.1128/EC.4.10.1687-1696.2005
- A. Sundaram, and C.M. Grant, "Oxidant-specific regulation of protein synthesis in Candida albicans", Fungal Genetics and Biology, vol. 67, pp. 15-23, 2014. http://dx.doi.org/10.1016/j.fgb.2014.03.005
- G. Tripathi, C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A.J.P. Brown, "Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans.", The EMBO journal, 2002. http://www.ncbi.nlm.nih.gov/pubmed/12374745
- A. Sundaram, and C.M. Grant, "A single inhibitory upstream open reading frame (uORF) is sufficient to regulateCandida albicans GCN4translation in response to amino acid starvation conditions", RNA, vol. 20, pp. 559-567, 2014. http://dx.doi.org/10.1261/rna.042267.113
- X. Wang, "Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo", The EMBO Journal, vol. 20, pp. 4349-4359, 2001. http://dx.doi.org/10.1093/emboj/20.16.4349
- L.K. Palmer, S.L. Rannels, S.R. Kimball, L.S. Jefferson, and R.L. Keil, "Inhibition of mammalian translation initiation by volatile anesthetics", American Journal of Physiology-Endocrinology and Metabolism, vol. 290, pp. E1267-E1275, 2006. http://dx.doi.org/10.1152/ajpendo.00463.2005
- L.K. Palmer, J.L. Shoemaker, B.A. Baptiste, D. Wolfe, and R.L. Keil, "Inhibition of Translation Initiation by Volatile Anesthetics Involves Nutrient-sensitive GCN-independent and -dependent Processes in Yeast", Molecular Biology of the Cell, vol. 16, pp. 3727-3739, 2005. http://dx.doi.org/10.1091/mbc.E05-02-0127
- D.A. Hogan, "Quorum Sensing: Alcohols in a Social Situation", Current Biology, vol. 16, pp. R457-R458, 2006. http://dx.doi.org/10.1016/j.cub.2006.05.035
- M.P. Ashe, "A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols", The EMBO Journal, vol. 20, pp. 6464-6474, 2001. http://dx.doi.org/10.1093/emboj/20.22.6464
- E.J. Taylor, S.G. Campbell, C.D. Griffiths, P.J. Reid, J.W. Slaven, R.J. Harrison, P.F. Sims, G.D. Pavitt, D. Delneri, and M.P. Ashe, "Fusel Alcohols Regulate Translation Initiation by Inhibiting eIF2B to Reduce Ternary Complex in a Mechanism That May Involve Altering the Integrity and Dynamics of the eIF2B Body", Molecular Biology of the Cell, vol. 21, pp. 2202-2216, 2010. http://dx.doi.org/10.1091/mbc.E09-11-0962
- N.E. Egbe, C.M. Paget, H. Wang, and M.P. Ashe, "Alcohols inhibit translation to regulate morphogenesis in C. albicans", Fungal Genetics and Biology, vol. 77, pp. 50-60, 2015. http://dx.doi.org/10.1016/j.fgb.2015.03.008
- J.D. Richter, and N. Sonenberg, "Regulation of cap-dependent translation by eIF4E inhibitory proteins", Nature, vol. 433, pp. 477-480, 2005. http://dx.doi.org/10.1038/nature03205
- M.L. Langford, S. Hasim, K.W. Nickerson, and A.L. Atkin, "Activity and Toxicity of Farnesol towards Candida albicans Are Dependent on Growth Conditions", Antimicrobial Agents and Chemotherapy, vol. 54, pp. 940-942, 2010. http://dx.doi.org/10.1128/AAC.01214-09
- S. Ibrahimo, L.E.A. Holmes, and M.P. Ashe, "Regulation of translation initiation by the yeast eIF4E binding proteins is required for the pseudohyphal response", Yeast, vol. 23, pp. 1075-1088, 2006. http://dx.doi.org/10.1002/yea.1415
- W.A. Fonzi, and M.Y. Irwin, "Isogenic strain construction and gene mapping in Candida albicans.", Genetics, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8349105
- J.F. Staab, and P. Sundstrom, "URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes.", Trends in microbiology, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12598128
- R. Shchepin, J.M. Hornby, E. Burger, T. Niessen, P. Dussault, and K.W. Nickerson, "Quorum sensing in Candida albicans: probing farnesol's mode of action with 40 natural and synthetic farnesol analogs.", Chemistry & biology, 2003. http://www.ncbi.nlm.nih.gov/pubmed/12954333
- J. Berman, "Morphogenesis and cell cycle progression in Candida albicans", Current Opinion in Microbiology, vol. 9, pp. 595-601, 2006. http://dx.doi.org/10.1016/j.mib.2006.10.007
- M.C. Lorenz, N.S. Cutler, and J. Heitman, "Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae.", Molecular biology of the cell, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10637301
- P. Sudbery, N. Gow, and J. Berman, "The distinct morphogenic states of Candida albicans", Trends in Microbiology, vol. 12, pp. 317-324, 2004. http://dx.doi.org/10.1016/j.tim.2004.05.008
- F.F. Ogletree, A.T. Abdelal, and D.G. Ahearn, "Germ-tube formation by atypical strains of Candida albicans.", Antonie van Leeuwenhoek, 1978. http://www.ncbi.nlm.nih.gov/pubmed/350146
- S.G. Campbell, N.P. Hoyle, and M.P. Ashe, "Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body", The Journal of Cell Biology, vol. 170, pp. 925-934, 2005. http://dx.doi.org/10.1083/jcb.200503162
- C.M. Browne, P. Samir, J.S. Fites, S.A. Villarreal, and A.J. Link, "The Yeast Eukaryotic Translation Initiation Factor 2B Translation Initiation Complex Interacts with the Fatty Acid Synthesis Enzyme YBR159W and Endoplasmic Reticulum Membranes", Molecular and Cellular Biology, vol. 33, pp. 1041-1056, 2013. http://dx.doi.org/10.1128/MCB.00811-12
- C. Noree, B.K. Sato, R.M. Broyer, and J.E. Wilhelm, "Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster", Journal of Cell Biology, vol. 190, pp. 541-551, 2010. http://dx.doi.org/10.1083/jcb.201003001
- K.H. Nielsen, B. Szamecz, L. Valášek, A. Jivotovskaya, B. Shin, and A.G. Hinnebusch, "Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control", The EMBO Journal, vol. 23, pp. 1166-1177, 2004. http://dx.doi.org/10.1038/sj.emboj.7600116
- L.M. Castelli, J. Lui, S.G. Campbell, W. Rowe, L.A.H. Zeef, L.E.A. Holmes, N.P. Hoyle, J. Bone, J.N. Selley, P.F.G. Sims, and M.P. Ashe, "Glucose depletion inhibits translation initiation via eIF4A loss and subsequent 48S preinitiation complex accumulation, while the pentose phosphate pathway is coordinately up-regulated", Molecular Biology of the Cell, vol. 22, pp. 3379-3393, 2011. http://dx.doi.org/10.1091/mbc.E11-02-0153
- H. Chen, and G.R. Fink, "Feedback control of morphogenesis in fungi by aromatic alcohols", Genes & Development, vol. 20, pp. 1150-1161, 2006. http://dx.doi.org/10.1101/gad.1411806
- J.R. Dickinson, "Fuse1 alcohols induce hyphal-like extensions and pseudohyphal formationin yeasts", Microbiology, vol. 142, pp. 1391-1397, 1996. http://dx.doi.org/10.1099/13500872-142-6-1391
- R.J. Rolfes, and A.G. Hinnebusch, "Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2.", Molecular and cellular biology, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8336737
- H. Kubota, T. Obata, K. Ota, T. Sasaki, and T. Ito, "Rapamycin-induced Translational Derepression of GCN4 mRNA Involves a Novel Mechanism for Activation of the eIF2α Kinase GCN2", Journal of Biological Chemistry, vol. 278, pp. 20457-20460, 2003. http://dx.doi.org/10.1074/jbc.C300133200
- T. Eom, V. Berardi, J. Zhong, G. Risuleo, and H. Tiedge, "Dual Nature of Translational Control by Regulatory BC RNAs", Molecular and Cellular Biology, vol. 31, pp. 4538-4549, 2011. http://dx.doi.org/10.1128/MCB.05885-11
- A. Cruz-Migoni, G.M. Hautbergue, P.J. Artymiuk, P.J. Baker, M. Bokori-Brown, C. Chang, M.J. Dickman, A. Essex-Lopresti, S.V. Harding, N.M. Mahadi, L.E. Marshall, G.W. Mobbs, R. Mohamed, S. Nathan, S.A. Ngugi, C. Ong, W.F. Ooi, L.J. Partridge, H.L. Phillips, M.F. Raih, S. Ruzheinikov, M. Sarkar-Tyson, S.E. Sedelnikova, S.J. Smither, P. Tan, R.W. Titball, S.A. Wilson, and D.W. Rice, "A Burkholderia pseudomallei Toxin Inhibits Helicase Activity of Translation Factor eIF4A", Science, vol. 334, pp. 821-824, 2011. http://dx.doi.org/10.1126/science.1211915
- H.A. Meijer, Y.W. Kong, W.T. Lu, A. Wilczynska, R.V. Spriggs, S.W. Robinson, J.D. Godfrey, A.E. Willis, and M. Bushell, "Translational Repression and eIF4A2 Activity Are Critical for MicroRNA-Mediated Gene Regulation", Science, vol. 340, pp. 82-85, 2013. http://dx.doi.org/10.1126/science.1231197
- S. Ghosh, B.W. Kebaara, A.L. Atkin, and K.W. Nickerson, "Regulation of Aromatic Alcohol Production in Candida albicans", Applied and Environmental Microbiology, vol. 74, pp. 7211-7218, 2008. http://dx.doi.org/10.1128/AEM.01614-08
- K. Weber, R. Sohr, B. Schulz, M. Fleischhacker, and M. Ruhnke, "Secretion of E , E -Farnesol and Biofilm Formation in Eight Different Candida Species", Antimicrobial Agents and Chemotherapy, vol. 52, pp. 1859-1861, 2008. http://dx.doi.org/10.1128/AAC.01646-07
- J.B. Smirnova, J.N. Selley, F. Sanchez-Cabo, K. Carroll, A.A. Eddy, J.E.G. McCarthy, S.J. Hubbard, G.D. Pavitt, C.M. Grant, and M.P. Ashe, "Global Gene Expression Profiling Reveals Widespread yet Distinctive Translational Responses to Different Eukaryotic Translation Initiation Factor 2B-Targeting Stress Pathways", Molecular and Cellular Biology, vol. 25, pp. 9340-9349, 2005. http://dx.doi.org/10.1128/MCB.25.21.9340-9349.2005
- L.M. Castelli, D. Talavera, C.J. Kershaw, S.S. Mohammad-Qureshi, J.L. Costello, W. Rowe, P.F.G. Sims, C.M. Grant, S.J. Hubbard, M.P. Ashe, and G.D. Pavitt, "The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation", PLOS Genetics, vol. 11, pp. e1005233, 2015. http://dx.doi.org/10.1371/journal.pgen.1005233
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We thank A. Brown (Aberdeen University), C. Grant (Uni-versity of Manchester (UoM)) and M. Pool (UoM) for strains and antibodies. We thank P. March and S. Mardsen for their help with the microscopy. The Bioimaging Facility microscopes used rely on grants by the Biotechnology and Biological Sciences Research Council (BBSRC), the Wellcome Trust and a UoM Strategic Fund. We are grateful to David Knight and Emma-Jane Keevil in the Biomolecular Analysis Core Facility, The University of Manchester for their help with mass spectrometry analysis. NEE was funded by the Tertiary education trust fund Nigeria, TD was funded by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)- science without borders program, CMP was funded by a BBSRC project grant (BB/K002767/1) and LMC was funded by a BBSRC LoLa grant (BB/G012571/1).
COPYRIGHT
© 2017

Farnesol inhibits translation to limit growth and filamentation in C. albicans and S. cerevisiae by Egbe is licensed under a Creative Commons Attribution 4.0 International License.