Microreviews:
Microbial Cell, Vol. 1, No. 1, pp. 45 - 47; doi: 10.15698/mic2014.01.125
Identifying the assembly pathway of cyanophage inside the marine bacterium using electron cryo-tomography
1 National Center for Macromolecular Imaging, Verna and Marrs Mclean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030 USA
2 Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139 USA
Keywords: electron cryo-tomography; Zernike phase optics; marine cyanobacterium; phage assembly intermediates; transient biological processes; 3-D structures.
Received originally: 09/12/2013 Accepted: 12/12/2013
Published: 06/01/2014
Correspondence:
Dr. Wah Chiu, National Center for Macromolecular Imaging, Verna and Marrs Mclean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX 77030 USA wah@bcm.edu
Conflict of interest statement: The authors declare no competing financial interests.
Please cite this article as: Wei Dai, Michael F. Schmid, Jonathan A. King Wah Chiu (2014). Identifying the assembly pathway of cyanophage inside the marine bacterium using electron cryo-tomography. Microbial Cell 1(1): 45-47.
Advances in electron cryo-tomography open up a new avenue to visualize the 3-D internal structure of a single bacterium before and after its infection by bacteriophages in its native environment, without using chemical fixatives, fluorescent dyes or negative stains. Such direct observation reveals the presence of assembly intermediates of the bacteriophage and thus allows us to map out the maturation pathway of the bacteriophage inside its host.
For every bacterium, there are associated bacterial viruses. The virus has dual effects on its bacterial host: destroying the bacteria through virus multiplication inside them leading to cell lysis, and enriching the gene pool of its host by incorporating host genes in its genome and transferring them among hosts. The processes of virus multiplication and lateral gene transfer have been critical areas of research in bacteriology and phage biology since the beginning of the molecular biology era. Through mutagenesis of the bacteriophage combined with biochemical analysis of the bacteria during the course of phage morphogenesis, it has been possible to dissect the pathway of the maturation process of many enteric bacteriophages within their hosts. However, such a discovery process requires tedious and laborious work in making many mutants, analyzing the protein profiles, and examining the cell structures by conventional electron microscopy of plastic embedded cell sections.
—
In electron cryo-tomography, the native structure of the bacteria is preserved in its physiological conditions for electron microscopic examination. Images of a frozen hydrated bacterium are recorded one at a time at different tilts, similar to a medical CT scan. These images can then be compiled to form a 3-D density volume where each voxel has a density value corresponding to the mass density of the biological contents of the bacterium. We applied this approach to study a Synechococcus cyanobacterium infected by Syn5 bacteriophage. Synechococcus WH8109 cell is a marine photosynthetic bacterium distributed throughout the world’s oceans. Marine bacteria play a critical role in the balance of the biosphere by absorbing carbon dioxide with the help of sunlight to generate biomass. It has been estimated that over 30% of carbon dioxide in the atmosphere are fixed by the marine bacteria through photosynthesis. Yet, very little is known about the pathogenesis of marine bacteria under the attack of cyanophages.
—
The Synechococcus WH8109 has a thickness of ~0.7 µm, which is thin enough to allow the 200 keV electrons to penetrate the cell in its entirety. The electron microscope used in our study was equipped with a special type of electron optics, in which the scattered electrons undergo a 90 degree phase shift. This is equivalent to the well-known light optics called Zernike phase contrast microscopy. The implementation of such optics in an electron microscope is relatively straightforward and involves placing a phase plate at the objective lens’ focal plane. The phase plate is made of a thin 20-25 nm carbon film with a hole (~1 µm diameter) in the middle of the carbon film. The electron images recorded this way yield a contrast enhancement of a factor of 3-5 relative to conventional electron optics. Though the physics principle is simple, the practice of this methodology to record high quality images is technically demanding because the phase plate is readily subject to deterioration under electron irradiation. Nevertheless, we were able to generate ~100 tilt series of Synechococcus cells infected by Syn5 bacteriophages, which allowed us to dissect the assembly pathway of Syn5 bacteriophage in its host bacteria.
—
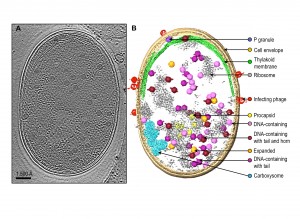
Since the infection process of the cell population by Syn5 bacteriophages is not tightly synchronized, our tomograms represent snapshots of the bacterium at different pathological states corresponding to different times post phage infection. Individual tomograms have remarkable contrast, such that many subcellular features can be visualized without ambiguity (Fig. 1). These features include the cell membrane, thylakoid membrane, ribosomes, carboxysome, bacteriophages at different stages of infection on the cell surface, and bacteriophage-like particles inside the cell with different shapes, sizes and densities. Data of this type certainly are unique among all imaging techniques because we are observing the biomolecules in the cells. This is distinct from the commonly used fluorescent optical microscopy, where only the labelled entity is identifiable and the molecular structure is completely obscure and invisible.
—
A challenge with our images is the need to manually annotate each of the observed molecular features. There is not yet a computational method to automatically pick out all the aforementioned subcellular features of interest. Human visual inspection and manual segmentation is the current practice to identify the salient features of interest (Fig. 1B). Generally, it takes ~ 40 hours of human effort to annotate apparently recognizable features in a single bacterial cell tomogram. Powerful computational tools are needed to extract the subcellular features of various types for analysis of their presence, relative abundance at different stages of the cells, their locations in the cell and their molecular structures.
Because the bacteriophages are relatively large (59-66 nm), they are easily detectable, and each phage-like particle can have its 3-D volume extracted for advanced computational analysis developed in our laboratory. Through this computational analysis, we were able to identify five different types of phage-like particles, which are interpreted as: procapsids with scaffolding protein inside the particle, expanded phages with terminase at one of the 12 vertices, DNA-containing phages, phages with fully packed DNA and a tail at one vertex, and mature phages with a tail at one vertex and a horn at the opposite vertex. Figure 1 shows an example of a slice from a tomogram of a phage-infected bacterium, and locations of different types of phages in the cytosol. The gallery of these phage structures averaged from multiple tomograms has taught us several things about the phage assembly process that were not fully known previously:
—
(1) Syn5 and other enteric phages are assembled with the assistance of scaffolding proteins that reside within the interior chamber of the particle. When the DNA is packaged into the procapsid, the scaffolding is believed to exit via the opening of the hexons with a concurrent conformational switch of the coat protein leading to the particle expansion. It is not known how soon the scaffolding proteins exit upon the initiation of the DNA packaging. The existence of expanded particles, which do not have extensive internal mass density, suggests that the scaffolding proteins must have exited the procapsid before the completion of DNA packaging. The small number of detectable particle of this type hints at their transient existence.
—
(2) the detection of a density at one vertex in the expanded particle is attributable to the terminase, which has not been seen in biochemically purified phage particles of any sort. This also suggests that the lifetime of this state is short (i.e. that DNA packaging is a relatively rapid event).
—
(3) the existence of the particles with tail alone and with tail and horn together indicates that the horn is added to the phage after the tail is assembled. This answers a frequently asked question for this phage about the biogenesis of the horn density located at the opposite vertex of the tail assembly.
—
This study illustrates that electron cryo-tomography is an approach whereby one can capture directly structural snapshots of transient phage assembly intermediates during maturation process. Such analysis can be generalizable not only to human viruses in human cells but also various molecular machines undergoing biological processes.
ACKNOWLEDGMENTS
This research has been supported by NIH grants (P41GM103832, PN2EY016525 and R56AI075208) and the Robert Welch Foundation (Q1242).
COPYRIGHT
© 2014

Identifying the assembly pathway of cyanophage inside the marine bacterium using electron cryo-tomography by Wei Dai et al. is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.