Back to article: Molecular signature of the imprintosome complex at the mating-type locus in fission yeast
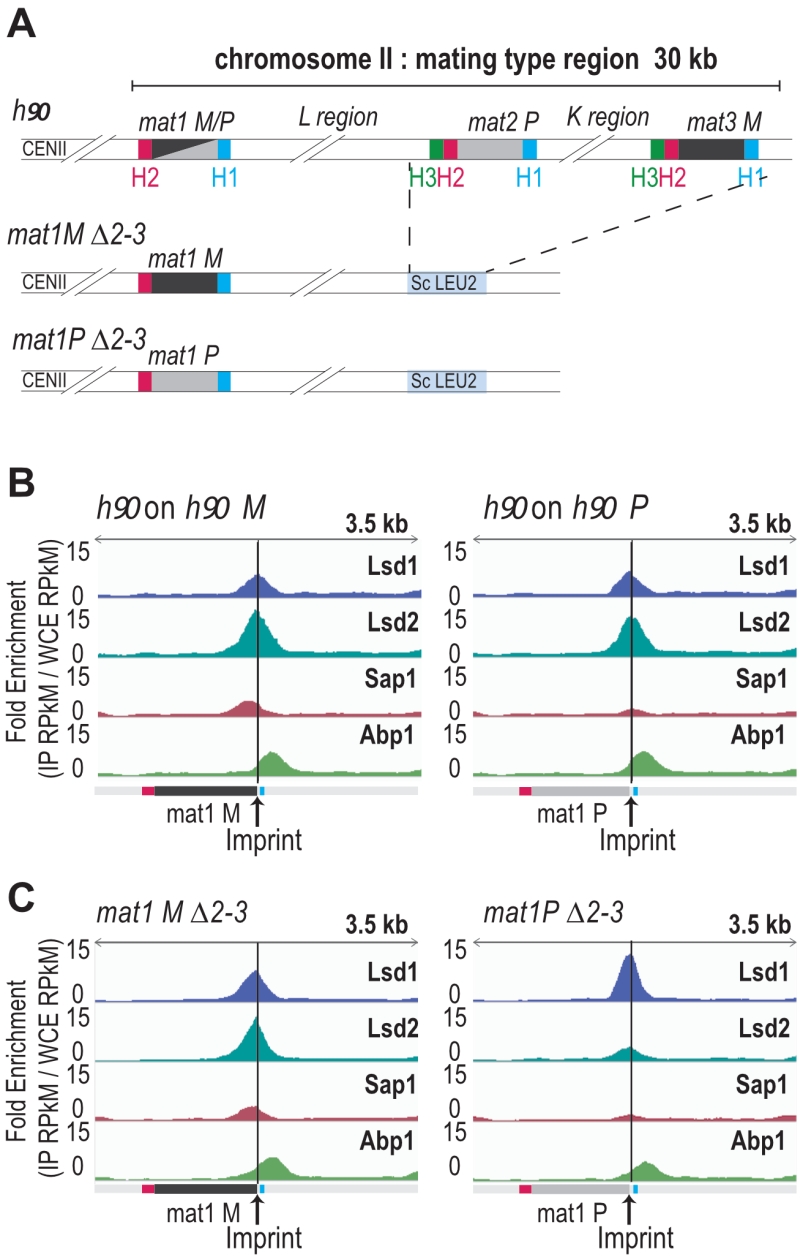
FIGURE 3: Abp1, Lsd1/2 and Sap1 are recruited at mat1. (A) Schematic view of the strains used. The stable mat1M ∆2-3 and mat1P ∆2-3 strains as well as the wild type h90 strain are drawn. (B) On the left: distribution of normalized enrichments of the Abp1, Lsd1, Lsd2, and Sap1 ChIPs on the h90 background (IP RPkM / WCE RPkM). The sequence used for the alignment is a 44-kb region that contains the MT region with the M allele at mat1: h90 M. Only mat1M is shown. On the right: same as at the left, with the h90 P sequence used for the alignment. Only mat1P is shown. (C) Same as in (B) with the mat1M ∆2-3 background on the left and the mat1P ∆2-3 background on the right. The sequence used for the alignments is a 10-kb region containing mat1M (for the left panel) or mat1P (for the right panel).( B-C) the imprint site is indicated as well as the H2 (pink) and H1 (blue) homology boxes.