Microreviews:
Microbial Cell, Vol. 1, No. 1, pp. 43 - 44; doi: 10.15698/mic2014.01.123
A novel mechanism involved in the coupling of mitochondrial biogenesis to oxidative phosphorylation
1 Centre de Génétique Moléculaire, Université Paris-Sud, avenue de la Terrasse, 91190 Gif sur Yvette, France
2 University Bordeaux, Institut de Biochimie et Génétique Cellulaires, UMR 5095, 33000 Bordeaux, France
3 CNRS, Institut de Biochimie et Génétique Cellulaires, UMR 5095, 33000 Bordeaux, France
Keywords: yeast, mitochondrial-disease model, AAA protein Bcs1, OXPHOS complex III, ATP synthase
Received originally: 28/11/2013 Received in revised form: 06/12/2013
Accepted: 06/12/2013
Published: 06/01/2014
Correspondence:
G. Dujardin, Centre de Génétique Moléculaire, Bâtiment 26, 1 avenue de la Terrasse, 91190 Gif sur Yvette, France dujardin@cgm.cnrs-gif.fr
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Jelena Ostojić, Jean-Paul di Rago, Geneviève Dujardin (2014). A novel mechanism involved in the coupling of mitochondrial biogenesis to oxidative phosphorylation. Microbial Cell 1(1): 43-44.
Mitochondria are essential organelles that are central to a multitude of cellular processes, including oxidative phosphorylation (OXPHOS), which produces most of the ATP in animal cells. Thus it is important to understand not only the mechanisms and biogenesis of this energy production machinery but also how it is regulated in both physiological and pathological contexts. A recent study by Ostojić et al. [Cell Metabolism (2013) 18, 567-577] has uncovered a regulatory loop by which the biogenesis of a major enzyme of the OXPHOS pathway, the respiratory complex III, is coupled to the energy producing activity of the mitochondria.
This study was performed in the yeast Saccharomyces cerevisiae, an organism particularly well suited to the analysis of mitochondrial functions. As a facultative aerobe, it can grow on fermentable sugars (e.g. glucose) and produce cellular energy (ATP) in the cytosol through substrate-level phosphorylation. Alternatively, yeast can synthesize ATP in mitochondria by the OXPHOS pathway. Because of this metabolic duality, yeast can tolerate mutations that inactivate oxidative phosphorylation as long as fermentable sugars are present in the growth medium. However, such mutants will be unable to grow on non-fermentable substrates that require functional mitochondria to be metabolized (e.g. glycerol, ethanol or lactate). These features provide a simple in vivo phenotypic readout of mitochondrial dysfunction that has been instrumental in the isolation of hundreds of yeast genes encoding mitochondrial proteins. Interestingly, 60% of these genes have human orthologs, many of which have been implicated in genetic diseases. As an easily tractable experimental system, where both the nuclear and mitochondrial genomes can be modified almost at will, yeast permits the modeling of many of the pathogenic mutations responsible for human mitochondrial diseases, allowing a better definition of their cellular consequences and helping in the search for potential rescue mechanisms.
—
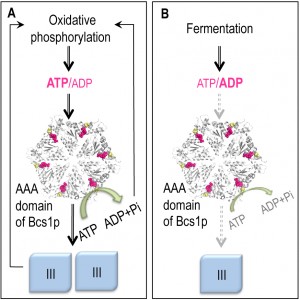
Ostojić et al. (2013) took advantage of the yeast system to further define the function of mitochondrial Bcs1p, the yeast homologue of human BCS1, which has been implicated in human mitochondrial disorders (e.g. Björnstad and Gracile syndromes). Bcs1 proteins belong to the large AAA-protein family (ATPase Associated with diverse cellular Activities) characterized by the presence of a highly conserved AAA region involved in ATP binding and hydrolysis. These proteins drive ATP-dependent dissociation, unfolding, or folding of nucleic acids and proteins. In mitochondria, they play a central role in the biogenesis and quality control of proteins. Bcs1 proteins are involved in the assembly of the respiratory complex III, one of the five complexes of the OXPHOS pathway. Complex III consists of 11 or 10 different subunits in mammals and yeast respectively, three of which are catalytic: cytochrome b, cytochrome c1 and the Rieske-FeS protein Rip1 (human UQCRFS1). Complex III is built through a sequential assembly of its subunits, the last step being the incorporation of Rip1, which is dependent on the hydrolysis of Bcs1-bound ATP.
—
A striking feature of Bcs1 deficiencies is the accumulation of an inactive pre-complex III lacking Rip1; this was also observed in our study in yeast strains carrying specific mutations in the AAA domain. These mutations impair the chaperon activity of yeast Bcs1p because they prevent Bcs1p from efficiently hydrolyzing ATP. We have isolated and characterized genetic suppressors able to restore Rip1p assembly in these mutants. Unexpectedly, the suppressor mutations mainly target the mitochondrial ATP synthase and lead to a strong decrease in the ATP hydrolytic activity while maintaining a sufficient level of ATP synthesis to sustain respiratory growth. We reasoned that by reducing the ATP hydrolytic activity of ATP synthase, the suppressors increase the concentration of ATP in the organelle and thereby enable the mutated Bcs1p to get a sufficient ATP hydrolysis activity to recover chaperon function. This hypothesis was supported by in vitro assays showing that the mutated protein does have a lower capacity to hydrolyze ATP compared to the wild type Bcs1p and that this difference disappears at higher ATP concentrations.
In respiring yeast cells, where large amounts of complex III are required, ATP is mostly synthesized in mitochondria and the rest of the cell is supplied by exchange against cytosolic ADP via the ADP/ATP translocase of the inner mitochondrial membrane. In fermenting cells, where there is no need to produce large amounts of complex III, ATP is mostly produced in the cytosol and imported into mitochondria using the same ADP/ATP translocase. The genetic interaction we found between Bcs1p and ATP synthase suggests that the AAA region of Bcs1p functions as a sensor of the ATP/ADP ratio in mitochondria to adjust the production of complex III according to the metabolic state of the cell.
–
The importance of ATP as a regulatory molecule is well established. However, so far, ATP has been found to modulate the activity of enzymes. Our study of Bcs1p reveals that ATP can be also used to influence the production of an enzyme by regulating a chaperone specifically involved in its assembly. We are currently testing the possibility that other AAA-proteins involved in mitochondrial biogenesis are regulated in a similar manner.
–
Our study identifies the intra-mitochondrial adenine nucleotide pool as a potential target for improving the condition of patients suffering from defects in mitochondrial ATP-dependent systems like Bcs1p. In a previous study, we reported the isolation of genetic suppressors able to improve mitochondrial ATP production in yeast models of human diseases caused by ATP synthase defects. Interestingly, subjecting these mutant yeast strains to chemical screens has identified drugs having a similar suppressor activity in both yeast and human ATP synthase defective cells. Using the same yeast-based pharmacological approach, we have isolated several compounds able to efficiently improve the respiratory capacity of yeast models of Bcs1-based disorders. These molecules could help in the development of treatments for Bcs1-based disorders and possibly other diseases involving ATP-dependent systems with a role in mitochondrial biogenesis.
ACKNOWLEDGMENTS
We thank Drs C. Panozzo and CJ Herbert for critical read-ing of the manuscript. J.O. was supported by a fellowship from the French Ministery of Research and Technologies.
COPYRIGHT
© 2014

A novel mechanism involved in the coupling of mitochondrial biogenesis to oxidative phosphorylation by Jelena Ostojić et al. is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.