Reviews:
Microbial Cell, Vol. 1, No. 10, pp. 318 - 324; doi: 10.15698/mic2014.10.169
The dual role of cyclin C connects stress regulated gene expression to mitochondrial dynamics
Department of Molecular Biology, Rowan University School of Osteopathic Medicine, Stratford NJ, USA.
Keywords: cyclin C, transcription, mediator, MAPK signal transduction pathway mitochondria, programmed cell death.
Abbreviations: PCD - programmed cell death, Cdks - cyclin dependent kinases, CWI - cell wall integrity.
Received originally: 02/05/2014 Received in revised form: 28/08/2014
Accepted: 02/09/2014
Published: 14/09/2014
Correspondence:
Katrina F. Cooper, Department of Molecular Biology, Rowan University, School of Osteopathic Medicine, Two Medical Center Drive; Stratford, NJ, 08055 USA cooperka@rowan.edu
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Randy Strich and Katrina F. Cooper (2014). The dual role of cyclin C connects stress regulated gene expression to mitochondrial dynamics. Microbial Cell 1(10): 318-324.
Abstract
Following exposure to cytotoxic agents, cellular damage is first recognized by a variety of sensor mechanisms. Thenceforth, the damage signal is transduced to the nucleus to install the correct gene expression program including the induction of genes whose products either detoxify destructive compounds or repair the damage they cause. Next, the stress signal is disseminated throughout the cell to effect the appropriate changes at organelles including the mitochondria. The mitochondria represent an important signaling platform for the stress response. An initial stress response of the mitochondria is extensive fragmentation. If the damage is prodigious, the mitochondria fragment (fission) and lose their outer membrane integrity leading to the release of pro-apoptotic factors necessary for programmed cell death (PCD) execution. As this complex biological process contains many moving parts, it must be exquisitely coordinated as the ultimate decision is life or death. The conserved C-type cyclin plays an important role in executing this molecular Rubicon by coupling changes in gene expression to mitochondrial fission and PCD. Cyclin C, along with its cyclin dependent kinase partner Cdk8, associates with the RNA polymerase holoenzyme to regulate transcription. In particular, cyclin C-Cdk8 repress many stress responsive genes. To relieve this repression, cyclin C is destroyed in cells exposed to pro-oxidants and other stressors. However, prior to its destruction, cyclin C, but not Cdk8, is released from its nuclear anchor (Med13), translocates from the nucleus to the cytoplasm where it interacts with the fission machinery and is both necessary and sufficient to induce extensive mitochondria fragmentation. Furthermore, cytoplasmic cyclin C promotes PCD indicating that it mediates both mitochondrial fission and cell death pathways. This review will summarize the role cyclin C plays in regulating stress-responsive transcription. In addition, we will detail this new function mediating mitochondrial fission and PCD. Although both these roles of cyclin C are conserved, this review will concentrate on cyclin C’s dual role in the budding yeast Saccharomyces cerevisiae.
ROLE 1: Cyclin C IS A TRANSCRIPTION FACTOR REPRESSING STRESS-RESPONSIVE GENES
Cyclin C-Cdk8 kinase is a part of the Mediator complex
The cyclin protein family was initially identified as promoters of cell cycle progression by binding and activating cyclin dependent kinases (Cdks). As indicated by their name, cyclins display a periodic expression pattern with their levels peaking at specific stages during mitotic cell division (reviewed in [1]). However, other cyclin-Cdk kinases were subsequently discovered that regulate transcription rather than cell cycle progression [2][3]. This group, cyclin C-Cdk8, cyclin H-Cdk7 and cyclin T-Cdk9 also share a commonality by associating with the RNA polymerase II machinery. Of these, cyclin C-Cdk8 share the most sequence conservation from yeast to man [4]. Structural analysis revealed specific determinants that promote cyclin C-Cdk8 interaction [5][6]. In addition, unlike other Cdks, Cdk8 does not require phosphorylation of the canonical T-loop for activation. Rather, the presence of an atypical acidic amino acid appears to have replaced this requirement [6].
–
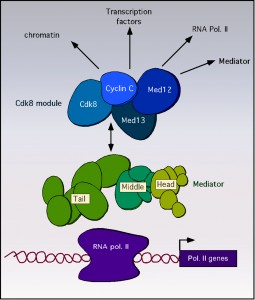
The cyclin C-Cdk8 kinase controls transcription though modification of the basal transcriptional machinery [7], chromatin [8][9] or transcription factors [10][11]. Recruitment of cyclin C-Cdk8 to promoters occurs through the Mediator, a large complex that plays a central role in modulating RNA polymerase II activity [12][13] (and reviewed in [14]). The mediator is comprised of 25-30 protein subunits that reside in four distinct domains termed head, middle, tail and the Cdk8 module (Figure 1). The Cdk8 module consists of cyclin C, Cdk8 and two additional proteins Med12 and Med13 [15][16]. This module is highly conserved and can be found either free [17] or associated with [12] the Mediator complex. The stoichiometry of the Cdk8 module is 1:1:1:1 and it associates with the core Mediator through the bridging function of Med13 [17][18].
The cyclin C-Cdk8 kinase primarily represses transcription of genes responding to environmental cues
Potential targets of cyclin C-Cdk8 that affect transcriptional control include other mediator components, transcription factors, chromatin and the RNA polymerase II itself (see [19] for review). Genetic studies in yeast first identified cyclin C (a.k.a. Ume3, Srb11, Ssn8) and Cdk8 (Ume5, Srb10, Ssn3) as negative transcriptional regulators of genes responding to environmental stimuli [20][21][22][23], (see [24] for review). Subsequent studies in yeast found that cyclin C-Cdk8 represses over 100 genes, many of which are considered stress response genes [25][26]. Although expression profiling indicates that cyclin C-Cdk8 plays largely a negative role in transcription, there are also reports of a positive role for this factor [10][27][28]. These positive and negative transcriptional regulatory roles of cyclin C-Cdk8 are dependent on specific promoter contexts (see [19][29] for recent reviews). Consistent with this observation, phenotypic studies have found that cyclin C-Cdk8 is required for processes that respond to a variety of external cues including meiotic development [30], pseudohyphal growth [11] and oxidative stress [25][31].
–
The stress-activated cell wall integrity MAPK pathway relieves cyclin C-Cdk8 repression
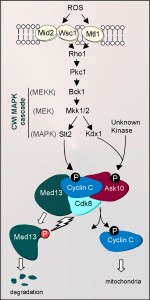
The other side of the coin with respect to cyclin C-Cdk8 repression is how this activity is removed to allow gene induction. Unlike cyclins that regulate the cell cycle, cyclin C levels do not vary significantly during the cell cycle in yeast or human cells [32][33]. However, in yeast, cyclin C-Cdk8 repression is relieved by cyclin destruction [32][34] through a Not4 ubiquitin ligase-dependent process [31]. In yeast, the cell wall integrity (CWI) signal transduction pathway responds to a variety of stresses including ROS [35], heat shock [36] and defects in CWI. The CWI pathway senses stress via cell-surface sensors (Wsc1-3, Mid2 and Mtl1, reviewed in [37]) that transmit the signal to a small G protein Rho1 (reviewed in [38]). Activated Rho1 stimulates protein kinase C (Pkc1, [39][40]) and the MAPK module composed of the MEK kinase Bck1, the redundant MEKs Mkk1 and Mkk2 [41], and the MAPK Slt2/Mpk1 [42], or its pseudokinase paralog Kdx1/Mlp1 ([43] and see Figure 2).
To affect transcription, the CWI pathway stimulates two well-characterized transcriptional activators Rlm1 and the heterodimeric factor Swi4-Swi6 (also termed SBF). Slt2 phosphorylates Rlm1 within its transcriptional activation domain to stimulate DNA binding [43][44]. Interestingly, although Slt2 phosphorylates Swi6 [45], a non-catalytic role for this kinase and Kdx1 in SBF-dependent activation has been described [46]. A non-catalytic role for transcriptional regulation is also observed during transcription elongation as well as initiation [47]. Importantly, both Slt2 and Kdx1 are activated by phosphorylation on their respective T-loop domains by Mkk1/Mkk2 [46]. In addition to stimulating transcription factors involved in stress gene induction, the CWI pathway is also responsible for mediating cyclin C destruction. For the cyclin C destruction pathway, oxidative stress is sensed by a complex combination of cell wall receptors (Wsc1, Mid2, Mtl1) whose activities are dictated by the level of oxidative damage [48]. For example, under low oxidative stress conditions, Mtl1, and either Wsc1 or Mid2, are required jointly to transmit the oxidative stress signal to initiate cyclin C destruction. However, when exposed to elevated oxidative stress, the activity of only one of these sensor groups is necessary to destroy cyclin C. In addition, N-glycosylation is important for Mtl1 function, as mutating the receptor residue (Asn42) or an enzyme required for synthesis of N-acetylglucosamine (Gfa1) reduces sensor activity [48]. These results provide a mechanism by which the cell is able to discern high- from low-level ROS damage to mediate cyclin C destruction.
–
Similar to activation of other transcription factors regulated by this pathway, the route the stress signal takes from Pkc1p to cyclin C is bifurcated at the MAP kinase step. The Slt2 MAPK directly phosphorylates cyclin C at Ser266 [49]. Eliminating this phosphorylation site prevents cyclin C proteolysis while a phosphomimetic mutation enhances its destruction kinetics. Conversely, the pseudokinase Kdx1 interacts with Ask10, a previously identified cyclin C associating factor [50]. Ask10 is required for efficient cyclin C destruction and is phosphorylated in response to H2O2 [49][50]. Interestingly, Ask10 phosphorylation requires the MEKs Mkk1 and Mkk2, the pseudokinase Kdx1, but not Slt2 [49][50]. Therefore, these results suggest the activity of another, unknown kinase in modifying Ask10 and controlling cyclin C destruction (Figure 2). Thus, cyclin C regulation is complex, as both Slt2 and Kdx1 are required for cyclin C destruction but do so through direct and indirect mechanisms, respectively. These findings emphasize that the molecular decision to destroy cyclin C is carefully regulated to prevent aberrant derepression of stress response genes.
ROLE 2: Cyclin C MEDIATES STRESS-INDUCED MITOCHONDRIA HYPER-FISSION
Stress-induced mitochondrial dynamics
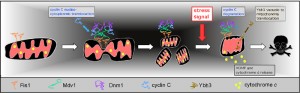
Similar to mammalian cells, yeast mitochondria serve as a signaling platform to both receive and send stress signals. Under normal growth conditions, mitochondria are usually observed in a more connected, reticular morphology (fusion) that enables maximum ATP production and the repair of mtDNA or damaged membranes by associating organelles. Conversely, mitochondrial fission allows isolation of defective organelles for removal via autophagy [51] or their efficient segregation at mitosis. However, in response to a variety of cytotoxic agents, the mitochondria undergo extensive fragmentation (Figure 3, see [52][53] for reviews). This hyper-fission is an important first step in the stress response pathway and observed in all organisms examined. Since the same machinery is utilized to execute normal mitochondrial division and hyper-fission, a stress-induced trigger for this process must be necessary.
Mitochondrial division machinery
Mitochondrial fission requires the conserved dynamin-like GTPase Dnm1. Dnm1 is recruited to the mitochondria by the outer membrane receptor Fis1 through one of two WD-40 adaptor proteins, Mdv1 [54][55] or Caf4 [56]. Functionally, Mdv1 acts as a nucleation factor for GTP bound Dnm1, recruiting it to the membrane via its C-terminal domain [57][58], and promoting Dnm1 to form spirals encircling the mitochondria [59]. GTP hydrolysis causes ring constriction that promotes scission [60]. X-ray structure analysis has revealed that Mdv1 dimerizes in an antiparallel coiled coil [61]. This coiled-coil domain also promotes assembly of Dnm1 oligomers, thus is important for filament formation. Caf4 also recruits Dnm1 to the mitochondria [56][62][63][64]. However, phenotypes associated with loss of Mdv1 function are more severe than in caf4∆ strains suggesting that Mdv1 plays a more essential role in fission [55][65].
–
Mitochondrial division machinery and PCD
As described above, exposure to adverse environmental conditions shifts the delicate balance between fission and fusion dramatically toward fission [52]. In yeast and higher eukaryotes, hyper-fission is proposed to be part of the process that leads to loss of mitochondrial membrane integrity and subsequent release of pro-apoptotic factors required for PCD execution. Many basic players in the PCD pathway have been conserved from yeast to humans such as caspases (reviewed in [66]), a Bcl-2 family member [67] and the nucleases Nuc1p and Aif1p that are responsible for chromatin cleavage (reviewed in [68][69]). However, some important mammalian PCD regulators (e.g., p53, SMAC) have not been identified to date in yeast. Consistent with a functional connection between hyper-fission and PCD, inactivating Dnm1 in yeast or its paralog in mammalian cells (Drp1) protects cells from PCD-inducing agents [70][71]. However, other reports have found that hyper-fission is not required for the release of all pro-apoptotic factors [72][73] suggesting that there may not be an absolute connection between the two processes.
–
Cytoplasmic localization of cyclin C directs stress-induced mitochondrial hyper-fission and programmed cell death
Several observations indicate that cyclin C directs stress-induced mitochondrial fission. First, before it is destroyed, cyclin C (but not Cdk8) translocates to the cytoplasm [31] where it associates with the mitochondria [74]. This latter study found that cyclin C is both necessary and sufficient for inducing extensive mitochondrial fragmentation. A mechanism to explain the role of cyclin C in mediating stress-induced fission is suggested by co-immunoprecipitation studies revealing that cyclin C associates with the fission machinery and is required for enhanced association of Dnm1 and Mdv1 in stressed cultures [74]. In addition, it was shown that cyclin C-Dnm1 association does not require mitochondrial association as the interaction was detected in fis1∆mutant strains. In addition, Dnm1 forms large, non-functional aggregates in cyclin C mutants. These findings indicate that cyclin C is required for normal assembly of functional Dnm1 filaments in stressed cells.
–
As indicated earlier, mitochondrial fission is associated with the initial stages of PCD. If there is causation between the two events, cyclin C would be predicted to be required for normal PCD execution. This is indeed the case as yeast strains lacking cyclin C are more resistant to ROS-induced programmed cell death [75]. However, ectopically inducing extensive fission by overexpressing cyclin C does not induce PCD [74] indicating that fission itself is not a commitment point for PCD execution in yeast. These findings suggest a two-step model for cyclin C-induced PCD. First, cyclin C localization to the mitochondria induces extensive fragmentation of this organelle. However, another cellular damage signal is required for the cell to commit to the cell death pathway. The nature of this signal is unknown at present. Interestingly, these results are different than those obtained with other PCD inducers that function at the mitochondria. For example, ectopically targeting p53 or bax to the mitochondria in non-stressed cells is sufficient to induce cell death [76][77]. Likewise, overexpression of the yeast BH3 domain protein (Ybh3) is also sufficient to induce cell death [66]. Therefore, cyclin C appears to represent a different class of regulator that is necessary and sufficient for hyper-fission but only necessary for efficient PCD.
–
Med13p anchors cyclin C in the nucleus in unstressed cells.
As stated above, in the nucleus, cyclin C-Cdk8 are components of a subcomplex of the mediator composed of Med12 and Med13. Recent studies have revealed that Med13 functions as an anchor protein that retains cyclin C in the nucleus in unstressed cultures [78]. Deleting MED13 allows constitutive cytoplasmic localization of cyclin C resulting in continuously fragmented mitochondria. The consequence of constant mitochondrial fragmentation is a loss of organelle function due to mtDNA deletions and a hypersensitivity to oxidative stress. To dissolve cyclin C-Med13 interaction, Med13 is subjected to ubiquitin-mediated destruction that is dependent on Cdk8 activity. In yeast strains expressing a kinase dead derivative of Cdk8, Med13 destruction is abrogated and cyclin C fails to leave the nucleus or nucleolus (Figure 2). Therefore, cyclin C release from the nucleus requires Slt2 phosphorylation and Med13 destruction, perhaps mediated by Cdk8 phosphorylation. These findings elaborate a complicated biochemical switch controlling cyclin C release that involves multiple signal transduction pathways and proteins that interact directly with cyclin C.
CONCLUSIONS AND FUTURE PERSPECTIVES
In response to stress, the cell must sense cellular damage, transmit this signal to the nucleus to alter gene expression programs, then finally alert the remainder of the cell to the damage. In the case of cyclin C, the cell has utilized re-localization strategies to solve this problem. Following the upstream and downstream components of this regulatory system, we observe an example of how the cell has been able to integrate multiple functions into a single protein. Being a single cell organism, yeast routinely encounters cytotoxic compounds that alter membrane fluidity, generating a signal that is transduced to the nucleus (Figure 3). In the nucleus, cyclin C phosphorylation induces its release from the nucleus resulting in derepression of stress response genes through inactivation of Cdk8p. In addition, its relocalization to the mitochondria represents an intracellular signal inducing extensive remodeling of the organelle that may result in cell death. This dual role for cyclin C allows the cell to couple gene expression with organelle dynamics to produce a coordinated response. In addition, both roles for cyclin C have been conserved in human cells (our unpublished results). These observations indicate that cyclin C-dependent control of transcription and mitochondrial dynamics is a very ancient process. It is clear that as metazoans developed, additional regulatory layers were applied to both transcriptional and PCD control. However, using yeast as a model has allowed the field to peel back the years to distill the basic regulatory and mechanistic threads of these diverse processes. Such knowledge will be important not only to provide a basic understanding of how these critical events are orchestrated, but also will help identify new players in these pathways that may provide new strategies to attack diseases such as cancer. For example, the ability to manipulate cyclin C localization affects cellular sensitivity to cytotoxic agents. Therefore, only a detailed knowledge of how this system works in normal cells will allow rational designs of potential therapeutics to be realized.
References
- A.W. Murray, "Recycling the Cell Cycle", Cell, vol. 116, pp. 221-234, 2004. http://dx.doi.org/10.1016/S0092-8674(03)01080-8
- B.D. Dynlacht, "Regulation of transcription by proteins that control the cell cycle", Nature, vol. 389, pp. 149-152, 1997. http://dx.doi.org/10.1038/38225
- D.B. Bregman, R.G. Pestell, and V.J. Kidd, "Cell cycle regulation and RNA polymerase II.", Frontiers in bioscience : a journal and virtual library, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10704151
- G. Lolli, "Structural dissection of cyclin dependent kinases regulation and protein recognition properties", Cell Cycle, vol. 9, pp. 1551-1561, 2010. http://dx.doi.org/10.4161/cc.9.8.11195
- S. Hoeppner, S. Baumli, and P. Cramer, "Structure of the Mediator Subunit Cyclin C and its Implications for CDK8 Function", Journal of Molecular Biology, vol. 350, pp. 833-842, 2005. http://dx.doi.org/10.1016/j.jmb.2005.05.041
- E. Schneider, J. Böttcher, M. Blaesse, L. Neumann, R. Huber, and K. Maskos, "The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder", Journal of Molecular Biology, vol. 412, pp. 251-266, 2011. http://dx.doi.org/10.1016/j.jmb.2011.07.020
- S. Akoulitchev, S. Chuikov, and D. Reinberg, "TFIIH is negatively regulated by cdk8-containing mediator complexes", Nature, vol. 407, pp. 102-106, 2000. http://dx.doi.org/10.1038/35024111
- M.T. Knuesel, K.D. Meyer, A.J. Donner, J.M. Espinosa, and D.J. Taatjes, "The Human CDK8 Subcomplex Is a Histone Kinase That Requires Med12 for Activity and Can Function Independently of Mediator", Molecular and Cellular Biology, vol. 29, pp. 650-661, 2009. http://dx.doi.org/10.1128/mcb.00993-08
- K.D. Meyer, A.J. Donner, M.T. Knuesel, A.G. York, J.M. Espinosa, and A.D.J. Taatjes, "Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3", The EMBO Journal, 2008. http://dx.doi.org/10.1038/emboj.2008.78
- M. Hirst, M.S. Kobor, N. Kuriakose, J. Greenblatt, and I. Sadowski, "GAL4 Is Regulated by the RNA Polymerase II Holoenzyme–Associated Cyclin-Dependent Protein Kinase SRB10/CDK8", Molecular Cell, vol. 3, pp. 673-678, 1999. http://dx.doi.org/10.1016/S1097-2765(00)80360-3
- C. Nelson, S. Goto, K. Lund, W. Hung, and I. Sadowski, "Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12", Nature, vol. 421, pp. 187-190, 2003. http://dx.doi.org/10.1038/nature01243
- R.C. Conaway, and J.W. Conaway, "Function and regulation of the Mediator complex", Current Opinion in Genetics & Development, vol. 21, pp. 225-230, 2011. http://dx.doi.org/10.1016/j.gde.2011.01.013
- S. BJORKLUND, and Y. KIM, "Mediator of transcriptional regulation", Trends in Biochemical Sciences, vol. 21, pp. 335-337, 1996. http://dx.doi.org/10.1016/S0968-0004(96)10051-7
- S.A. Ansari, and R.H. Morse, "Mechanisms of Mediator complex action in transcriptional activation", Cellular and Molecular Life Sciences, vol. 70, pp. 2743-2756, 2013. http://dx.doi.org/10.1007/s00018-013-1265-9
- T. Borggrefe, R. Davis, H. Erdjument-Bromage, P. Tempst, and R.D. Kornberg, "A Complex of the Srb8, -9, -10, and -11 Transcriptional Regulatory Proteins from Yeast", Journal of Biological Chemistry, vol. 277, pp. 44202-44207, 2002. http://dx.doi.org/10.1074/jbc.M207195200
- E. Larschan, and F. Winston, "The Saccharomyces cerevisiae Srb8-Srb11 Complex Functions with the SAGA Complex during Gal4-Activated Transcription", Molecular and Cellular Biology, vol. 25, pp. 114-123, 2005. http://dx.doi.org/10.1128/MCB.25.1.114-123.2005
- M.T. Knuesel, K.D. Meyer, C. Bernecky, and D.J. Taatjes, "The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function", Genes & Development, vol. 23, pp. 439-451, 2009. http://dx.doi.org/10.1101/gad.1767009
- K. Tsai, S. Sato, C. Tomomori-Sato, R.C. Conaway, J.W. Conaway, and F.J. Asturias, "A conserved Mediator–CDK8 kinase module association regulates Mediator–RNA polymerase II interaction", Nature Structural & Molecular Biology, vol. 20, pp. 611-619, 2013. http://dx.doi.org/10.1038/nsmb.2549
- J. Nemet, B. Jelicic, I. Rubelj, and M. Sopta, "The two faces of Cdk8, a positive/negative regulator of transcription", Biochimie, vol. 97, pp. 22-27, 2014. http://dx.doi.org/10.1016/j.biochi.2013.10.004
- S. Kuchin, P. Yeghiayan, and M. Carlson, "Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast.", Proceedings of the National Academy of Sciences, vol. 92, pp. 4006-4010, 1995. http://dx.doi.org/10.1073/pnas.92.9.4006
- R.T. Surosky, R. Strich, and R.E. Esposito, "The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose.", Molecular and cellular biology, 1994. http://www.ncbi.nlm.nih.gov/pubmed/8164691
- M. Carlson, B.C. Osmond, L. Neigeborn, and D. Botstein, "A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast.", Genetics, 1984. http://www.ncbi.nlm.nih.gov/pubmed/6373495
- R. Strich, M.R. Slater, and R.E. Esposito, "Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast.", Proceedings of the National Academy of Sciences of the United States of America, 1989. http://www.ncbi.nlm.nih.gov/pubmed/2690066
- M. Carlson, "GENETICS OF TRANSCRIPTIONAL REGULATION IN YEAST: Connections to the RNA Polymerase II CTD", Annual Review of Cell and Developmental Biology, vol. 13, pp. 1-23, 1997. http://dx.doi.org/10.1146/annurev.cellbio.13.1.1
- F.C. Holstege, E.G. Jennings, J.J. Wyrick, T.I. Lee, C.J. Hengartner, M.R. Green, T.R. Golub, E.S. Lander, and R.A. Young, "Dissecting the Regulatory Circuitry of a Eukaryotic Genome", Cell, vol. 95, pp. 717-728, 1998. http://dx.doi.org/10.1016/S0092-8674(00)81641-4
- J. van de Peppel, N. Kettelarij, H. van Bakel, T.T. Kockelkorn, D. van Leenen, and F.C. Holstege, "Mediator Expression Profiling Epistasis Reveals a Signal Transduction Pathway with Antagonistic Submodules and Highly Specific Downstream Targets", Molecular Cell, vol. 19, pp. 511-522, 2005. http://dx.doi.org/10.1016/j.molcel.2005.06.033
- K. Hirst, F. Fisher, P.C. McAndrew, and C.R. Goding, "The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal.", The EMBO journal, 1994. http://www.ncbi.nlm.nih.gov/pubmed/7957107
- O. Vincent, S. Kuchin, S. Hong, R. Townley, V.K. Vyas, and M. Carlson, "Interaction of the Srb10 Kinase with Sip4, a Transcriptional Activator of Gluconeogenic Genes in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 21, pp. 5790-5796, 2001. http://dx.doi.org/10.1128/MCB.21.17.5790-5796.2001
- W. Xu, and J. Ji, "Dysregulation of CDK8 and Cyclin C in tumorigenesis", Journal of Genetics and Genomics, vol. 38, pp. 439-452, 2011. http://dx.doi.org/10.1016/j.jgg.2011.09.002
- K.F. Cooper, and R. Strich, "Saccharomyces cerevisiae C-Type Cyclin Ume3p/Srb11p Is Required for Efficient Induction and Execution of Meiotic Development", Eukaryotic Cell, vol. 1, pp. 66-74, 2002. http://dx.doi.org/10.1128/EC.01.1.66-74.2002
- K.F. Cooper, M.S. Scarnati, E. Krasley, M.J. Mallory, C. Jin, M.J. Law, and R. Strich, "Oxidative-stress-induced nuclear to cytoplasmic relocalization is required for Not4-dependent cyclin C destruction", Journal of Cell Science, vol. 125, pp. 1015-1026, 2012. http://dx.doi.org/10.1242/jcs.096479
- K.F. Cooper, "Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p)", The EMBO Journal, vol. 16, pp. 4665-4675, 1997. http://dx.doi.org/10.1093/emboj/16.15.4665
- D.J. Lew, V. Dulić, and S.I. Reed, "Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast.", Cell, 1991. http://www.ncbi.nlm.nih.gov/pubmed/1833066
- K.F. Cooper, M.J. Mallory, and R. Strich, "Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome.", Molecular and cellular biology, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10207058
- F. Vilella, E. Herrero, J. Torres, and M.A. de la Torre-Ruiz, "Pkc1 and the Upstream Elements of the Cell Integrity Pathway in Saccharomyces cerevisiae, Rom2 and Mtl1, Are Required for Cellular Responses to Oxidative Stress", Journal of Biological Chemistry, vol. 280, pp. 9149-9159, 2005. http://dx.doi.org/10.1074/jbc.M411062200
- Y. Kamada, U.S. Jung, J. Piotrowski, and D.E. Levin, "The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response.", Genes & Development, vol. 9, pp. 1559-1571, 1995. http://dx.doi.org/10.1101/gad.9.13.1559
- A. Jendretzki, J. Wittland, S. Wilk, A. Straede, and J.J. Heinisch, "How do I begin? Sensing extracellular stress to maintain yeast cell wall integrity", European Journal of Cell Biology, vol. 90, pp. 740-744, 2011. http://dx.doi.org/10.1016/j.ejcb.2011.04.006
- D.E. Levin, "Regulation of Cell Wall Biogenesis in Saccharomyces cerevisiae: The Cell Wall Integrity Signaling Pathway", Genetics, vol. 189, pp. 1145-1175, 2011. http://dx.doi.org/10.1534/genetics.111.128264
- H. Nonaka, K. Tanaka, H. Hirano, T. Fujiwara, H. Kohno, M. Umikawa, A. Mino, and Y. Takai, "A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae.", The EMBO journal, 1995. http://www.ncbi.nlm.nih.gov/pubmed/8846785
- Y. Kamada, H. Qadota, C.P. Python, Y. Anraku, Y. Ohya, and D.E. Levin, "Activation of Yeast Protein Kinase C by Rho1 GTPase", Journal of Biological Chemistry, vol. 271, pp. 9193-9196, 1996. http://dx.doi.org/10.1074/jbc.271.16.9193
- K. Irie, M. Takase, K.S. Lee, D.E. Levin, H. Araki, K. Matsumoto, and Y. Oshima, "MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C.", Molecular and cellular biology, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8386320
- K.S. Lee, K. Irie, Y. Gotoh, Y. Watanabe, H. Araki, E. Nishida, K. Matsumoto, and D.E. Levin, "A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C.", Molecular and cellular biology, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8386319
- Y. Watanabe, G. Takaesu, M. Hagiwara, K. Irie, and K. Matsumoto, "Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway.", Molecular and cellular biology, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9111331
- U.S. Jung, A.K. Sobering, M.J. Romeo, and D.E. Levin, "Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase", Molecular Microbiology, vol. 46, pp. 781-789, 2002. http://dx.doi.org/10.1046/j.1365-2958.2002.03198.x
- K. Madden, Y. Sheu, K. Baetz, B. Andrews, and M. Snyder, "SBF Cell Cycle Regulator as a Target of the Yeast PKC-MAP Kinase Pathway", Science, vol. 275, pp. 1781-1784, 1997. http://dx.doi.org/10.1126/science.275.5307.1781
- K. Kim, A.W. Truman, and D.E. Levin, "Yeast Mpk1 Mitogen-Activated Protein Kinase Activates Transcription through Swi4/Swi6 by a Noncatalytic Mechanism That Requires Upstream Signal", Molecular and Cellular Biology, vol. 28, pp. 2579-2589, 2008. http://dx.doi.org/10.1128/MCB.01795-07
- K. Kim, and D. Levin, "Mpk1 MAPK Association with the Paf1 Complex Blocks Sen1-Mediated Premature Transcription Termination", Cell, vol. 144, pp. 745-756, 2011. http://dx.doi.org/10.1016/j.cell.2011.01.034
- C. Jin, A.V. Parshin, I. Daly, R. Strich, and K.F. Cooper, "The Cell Wall Sensors Mtl1, Wsc1, and Mid2 Are Required for Stress-Induced Nuclear to Cytoplasmic Translocation of Cyclin C and Programmed Cell Death in Yeast", Oxidative Medicine and Cellular Longevity, vol. 2013, pp. 1-15, 2013. http://dx.doi.org/10.1155/2013/320823
- C. Jin, R. Strich, and K.F. Cooper, "Slt2p phosphorylation induces cyclin C nuclear-to-cytoplasmic translocation in response to oxidative stress", Molecular Biology of the Cell, vol. 25, pp. 1396-1407, 2014. http://dx.doi.org/10.1091/mbc.E13-09-0550
- T.J. Cohen, K. Lee, L.H. Rutkowski, and R. Strich, "Ask10p Mediates the Oxidative Stress-Induced Destruction of the Saccharomyces cerevisiae C-Type Cyclin Ume3p/Srb11p", Eukaryotic Cell, vol. 2, pp. 962-970, 2003. http://dx.doi.org/10.1128/EC.2.5.962-970.2003
- G. Twig, A. Elorza, A.J.A. Molina, H. Mohamed, J.D. Wikstrom, G. Walzer, L. Stiles, S.E. Haigh, S. Katz, G. Las, J. Alroy, M. Wu, B.F. Py, J. Yuan, J.T. Deeney, B.E. Corkey, and O.S. Shirihai, "Fission and selective fusion govern mitochondrial segregation and elimination by autophagy", The EMBO Journal, vol. 27, pp. 433-446, 2008. http://dx.doi.org/10.1038/sj.emboj.7601963
- B. Westermann, "Mitochondrial fusion and fission in cell life and death", Nature Reviews Molecular Cell Biology, vol. 11, pp. 872-884, 2010. http://dx.doi.org/10.1038/nrm3013
- J.R. Friedman, and J. Nunnari, "Mitochondrial form and function", Nature, vol. 505, pp. 335-343, 2014. http://dx.doi.org/10.1038/nature12985
- A. Mozdy, J. McCaffery, and J. Shaw, "Dnm1p Gtpase-Mediated Mitochondrial Fission Is a Multi-Step Process Requiring the Novel Integral Membrane Component Fis1p", The Journal of Cell Biology, vol. 151, pp. 367-380, 2000. http://dx.doi.org/10.1083/jcb.151.2.367
- Q. Tieu, and J. Nunnari, "Mdv1p Is a Wd Repeat Protein That Interacts with the Dynamin-Related Gtpase, Dnm1p, to Trigger Mitochondrial Division", The Journal of Cell Biology, vol. 151, pp. 353-366, 2000. http://dx.doi.org/10.1083/jcb.151.2.353
- E.E. Griffin, J. Graumann, and D.C. Chan, "The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria", The Journal of Cell Biology, vol. 170, pp. 237-248, 2005. http://dx.doi.org/10.1083/jcb.200503148
- L.L. Lackner, J.S. Horner, and J. Nunnari, "Mechanistic Analysis of a Dynamin Effector", Science, vol. 325, pp. 874-877, 2009. http://dx.doi.org/10.1126/science.1176921
- S. Koirala, H.T. Bui, H.L. Schubert, D.M. Eckert, C.P. Hill, M.S. Kay, and J.M. Shaw, "Molecular architecture of a dynamin adaptor: implications for assembly of mitochondrial fission complexes", Journal of Cell Biology, vol. 191, pp. 1127-1139, 2010. http://dx.doi.org/10.1083/jcb.201005046
- E. Ingerman, E.M. Perkins, M. Marino, J.A. Mears, J.M. McCaffery, J.E. Hinshaw, and J. Nunnari, "Dnm1 forms spirals that are structurally tailored to fit mitochondria", The Journal of Cell Biology, vol. 170, pp. 1021-1027, 2005. http://dx.doi.org/10.1083/jcb.200506078
- J.A. Mears, L.L. Lackner, S. Fang, E. Ingerman, J. Nunnari, and J.E. Hinshaw, "Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission", Nature Structural & Molecular Biology, vol. 18, pp. 20-26, 2010. http://dx.doi.org/10.1038/nsmb.1949
- Y. Zhang, and D.C. Chan, "Structural basis for recruitment of mitochondrial fission complexes by Fis1", Proceedings of the National Academy of Sciences, vol. 104, pp. 18526-18530, 2007. http://dx.doi.org/10.1073/pnas.0706441104
- Q. Guo, S. Koirala, E.M. Perkins, J.M. McCaffery, and J.M. Shaw, "The Mitochondrial Fission Adaptors Caf4 and Mdv1 Are Not Functionally Equivalent", PLoS ONE, vol. 7, pp. e53523, 2012. http://dx.doi.org/10.1371/journal.pone.0053523
- A.M. Motley, G.P. Ward, and E.H. Hettema, "Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p", Journal of Cell Science, vol. 121, pp. 1633-1640, 2008. http://dx.doi.org/10.1242/jcs.026344
- A.C. Schauss, J. Bewersdorf, and S. Jakobs, "Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface", Journal of Cell Science, vol. 119, pp. 3098-3106, 2006. http://dx.doi.org/10.1242/jcs.03026
- K.L. Cerveny, J.M. McCaffery, and R.E. Jensen, "Division of Mitochondria Requires a NovelDNM1-interacting Protein, Net2p", Molecular Biology of the Cell, vol. 12, pp. 309-321, 2001. http://dx.doi.org/10.1091/mbc.12.2.309
- D. Wilkinson, and M. Ramsdale, "Proteases and caspase-like activity in the yeast Saccharomyces cerevisiae", Biochemical Society Transactions, vol. 39, pp. 1502-1508, 2011. http://dx.doi.org/10.1042/BST0391502
- S. Büttner, D. Ruli, F. Vögtle, L. Galluzzi, B. Moitzi, T. Eisenberg, O. Kepp, L. Habernig, D. Carmona-Gutierrez, P. Rockenfeller, P. Laun, M. Breitenbach, C. Khoury, K. Fröhlich, G. Rechberger, C. Meisinger, G. Kroemer, and F. Madeo, "A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis", The EMBO Journal, vol. 30, pp. 2779-2792, 2011. http://dx.doi.org/10.1038/emboj.2011.197
- D. Carmona-Gutierrez, T. Eisenberg, S. Büttner, C. Meisinger, G. Kroemer, and F. Madeo, "Apoptosis in yeast: triggers, pathways, subroutines", Cell Death & Differentiation, vol. 17, pp. 763-773, 2010. http://dx.doi.org/10.1038/cdd.2009.219
- K. Fröhlich, H. Fussi, and C. Ruckenstuhl, "Yeast apoptosis—From genes to pathways", Seminars in Cancer Biology, vol. 17, pp. 112-121, 2007. http://dx.doi.org/10.1016/j.semcancer.2006.11.006
- Y. Fannjiang, W. Cheng, S.J. Lee, B. Qi, J. Pevsner, J.M. McCaffery, R.B. Hill, G. Basañez, and J.M. Hardwick, "Mitochondrial fission proteins regulate programmed cell death in yeast", Genes & Development, vol. 18, pp. 2785-2797, 2004. http://dx.doi.org/10.1101/gad.1247904
- N. Ishihara, A. Jofuku, Y. Eura, and K. Mihara, "Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells", Biochemical and Biophysical Research Communications, vol. 301, pp. 891-898, 2003. http://dx.doi.org/10.1016/S0006-291X(03)00050-0
- D. Arnoult, A. Grodet, Y. Lee, J. Estaquier, and C. Blackstone, "Release of OPA1 during Apoptosis Participates in the Rapid and Complete Release of Cytochrome c and Subsequent Mitochondrial Fragmentation", Journal of Biological Chemistry, vol. 280, pp. 35742-35750, 2005. http://dx.doi.org/10.1074/jbc.M505970200
- P.A. Parone, D.I. James, S. Da Cruz, Y. Mattenberger, O. Donzé, F. Barja, and J. Martinou, "Inhibiting the Mitochondrial Fission Machinery Does Not Prevent Bax/Bak-Dependent Apoptosis", Molecular and Cellular Biology, vol. 26, pp. 7397-7408, 2006. http://dx.doi.org/10.1128/MCB.02282-05
- K. Cooper, S. Khakhina, S. Kim, and R. Strich, "Stress-Induced Nuclear-to-Cytoplasmic Translocation of Cyclin C Promotes Mitochondrial Fission in Yeast", Developmental Cell, vol. 28, pp. 161-173, 2014. http://dx.doi.org/10.1016/j.devcel.2013.12.009
- E. Krasley, K.F. Cooper, M.J. Mallory, R. Dunbrack, and R. Strich, "Regulation of the Oxidative Stress Response Through Slt2p-Dependent Destruction of Cyclin C in Saccharomyces cerevisiae", Genetics, vol. 172, pp. 1477-1486, 2006. http://dx.doi.org/10.1534/genetics.105.052266
- U.M. Moll, S. Wolff, D. Speidel, and W. Deppert, "Transcription-independent pro-apoptotic functions of p53", Current Opinion in Cell Biology, vol. 17, pp. 631-636, 2005. http://dx.doi.org/10.1016/j.ceb.2005.09.007
- M. Karbowski, K.L. Norris, M.M. Cleland, S. Jeong, and R.J. Youle, "Role of Bax and Bak in mitochondrial morphogenesis", Nature, vol. 443, pp. 658-662, 2006. http://dx.doi.org/10.1038/nature05111
- S. Khakhina, K.F. Cooper, and R. Strich, "Med13p prevents mitochondrial fission and programmed cell death in yeast through nuclear retention of cyclin C", Molecular Biology of the Cell, vol. 25, pp. 2807-2816, 2014. http://dx.doi.org/10.1091/mbc.E14-05-0953
ACKNOWLEDGMENTS
We thank members of the Strich and Cooper laboratories for helpful comments. This review is based on work that is funded from grants to R.S. from the National Institutes of Health (RO1CA099003, RO1GM086788) and the WW Smith Charitable Trust (#CO604) to K.F.C.
COPYRIGHT
© 2014

The dual role of cyclin C connects stress regulated gene expression to mitochondrial dynamics by Randy Strich and Katrina F. Cooper is licensed under a Creative Commons Attribution 4.0 International License.