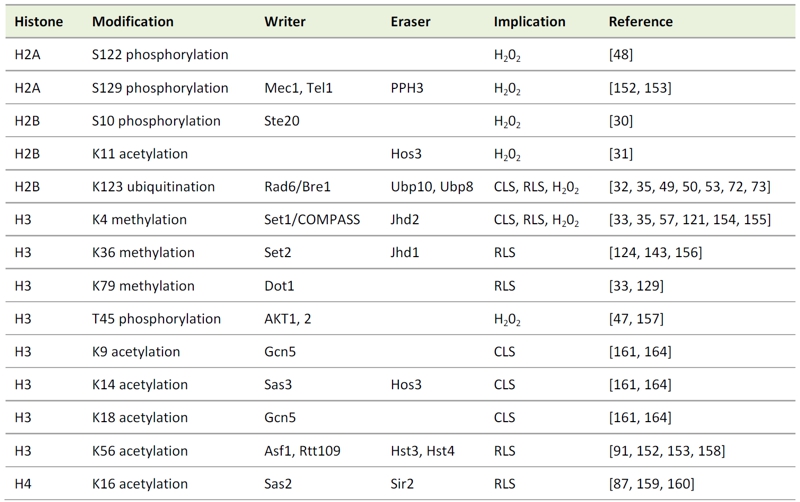
TABLE 1. Histone modifications involved in aging and apoptotic processes in yeast.
The implications listed are referring to the respective histone modifications. The listed writers and erasers may have impact on other histone modifications not related to cell death as well and the modifiers may have targets other than the histones, which may implicate them in other cell death pathways.
30. Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM (2000). Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem 275(13): 9390-9395. https://doi.org/10.1074/jbc.275.13.9390
31. Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, Allis CD (2003). Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell 113(4): 507-517. https://doi.org/10.1016/S0092-8674(03)00355-6
32. Ajiro K (2000). Histone H2B phosphorylation in mammalian apoptotic cells. An association with DNA fragmentation. J Biol Chem 275(1): 439-443. https://doi.org/10.1074/jbc.275.1.439
33. Fernandez-Capetillo O, Allis CD, Nussenzweig A (2004). Phosphorylation of histone H2B at DNA double-strand breaks. J Exp Med 199(12): 1671-1677. https://doi.org/10.1084/jem.20032247
35. Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001). Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8(2): 473-479. https://doi.org/10.1016/S1097-2765(01)00301-X
47. Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL (2007). H2B Ubiquitylation Acts as a Barrier to Ctk1 Nucleosomal Recruitment Prior to Removal by Ubp8 within a SAGA-Related Complex. Mol Cell 27(2): 275-288. https://doi.org/10.1016/j.molcel.2007.01.035
48. Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD (2005). Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Molecular and cellular biology 25(2): 637-651. https://doi.org/10.1128/MCB.25.2.637-651.2005
49. Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A (2002). Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. The Journal of biological chemistry 277(32): 28368-28371. https://doi.org/10.1074/jbc.C200348200
50. Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001). Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15(24): 3286-3295. https://doi.org/10.1101/gad.940201
53. Chandrasekharan MB, Huang F, Sun ZW (2009). Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A 106(39): 16686-16691. https://doi.org/10.1073/pnas.0907862106
57. Game JC, Williamson MS, Spicakova T, Brown M (2006). The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics 137(4): 1951-1968. https://doi.org/10.1534/genetics.106.057794
72. Lettre G, Kritikou EA, Jaeggi M, Calixto A, Fraser AG, Kamath RS, Ahringer J, Hengartner MO (2004). Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell death and differentiation 11(11): 1198-1203. https://doi.org/10.1038/sj.cdd.4401488
73. Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA, Chernikova DA, Kwok S, Brooks JD, Bailey SM, Game JC, Brown JM (2012). Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res 72(8): 2111-2119. https://doi.org/10.1158/0008-5472.CAN-11-2209
87. Xiao T, Shibata Y, Rao B, Laribee RN, O’Rourke R, Buck MJ, Greenblatt JF, Krogan NJ, Lieb JD, Strahl BD (2007). The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol Cell Biol 27(2): 721-731. https://doi.org/10.1128/MCB.01628-06
91. Ng HH, Robert F, Young RA, Struhl K (2003). Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11(3): 709-719. https://doi.org/10.1016/s1097-2765(03)00092-3
121. Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A (2010). Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev 24(6): 574-589. https://doi.org/10.1101/gad.1898410
124. Smeal T, Claus J, Kennedy B, Cole F, Guarente L (1996). Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84(4): 633-642. http://www.ncbi.nlm.nih.gov/pubmed/?term=8598049
129. Smith JS, Boeke JD (1997). An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11(2): 241-254. http://www.ncbi.nlm.nih.gov/pubmed/?term=9009206
143. Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK (2008). Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134(2): 231-243. https://doi.org/10.1016/j.cell.2008.06.035
152. Downs JA, Lowndes NF, Jackson SP (2000). A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408(6815): 1001-1004. https://doi.org/10.1038/35050000
153. Keogh MC, Kim JA, Downey M, Fillingham J, Chowdhury D, Harrison JC, Onishi M, Datta N, Galicia S, Emili A, Lieberman J, Shen X, Buratowski S, Haber JE, Durocher D, Greenblatt JF, Krogan NJ (2006). A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439(7075): 497-501. https://doi.org/10.1038/nature04384
154. Liang G, Klose RJ, Gardner KE, Zhang Y (2007). Yeast Jhd2p is a histone H3 Lys4 trimethyl demethylase. Nat Struct Mol Biol 14(3): 243-245. https://doi.org/10.1038/nsmb1204
155. Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A (2010). Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466(7304): 383-387. https://doi.org/10.1038/nature09195
156. Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, Freitas MA, Tsai MD (2007). Identification of histone demethylases in Saccharomyces cerevisiae. J Biol Chem 282(19): 14262-14271. https://doi.org/10.1074/jbc.M609900200
157. Lee JH, Kang BH, Jang H, Kim TW, Choi J, Kwak S, Han J, Cho EJ, Youn HD (2015). AKT phosphorylates H3-threonine 45 to facilitate termination of gene transcription in response to DNA damage. Nucleic Acids Res 43(9): 4505-4516. https://doi.org/10.1093/nar/gkv176
158. Hiraga S, Botsios S, Donaldson AD (2008). Histone H3 lysine 56 acetylation by Rtt109 is crucial for chromosome positioning. J Cell Biol 183(4): 641-651. https://doi.org/10.1083/jcb.200806065
159. Kozak ML, Chavez A, Dang W, Berger SL, Ashok A, Guo X, Johnson FB (2010). Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J 29(1): 158-170. https://doi.org/10.1038/emboj.2009.314
160. Vaquero A, Sternglanz R, Reinberg D (2007). NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 26(37): 5505-5520. https://doi.org/10.1038/sj.onc.1210617
161. Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Frohlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F (2009). Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11(11): 1305-1314. https://doi.org/10.1038/ncb1975
164. Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Kuttner V, Bhukel A, Marino G, Pietrocola F, Harger A, Zimmermann A, Moustafa T, Sprenger A, Jany E, Buttner S, Carmona-Gutierrez D, Ruckenstuhl C, Ring J, Reichelt W, Schimmel K, Leeb T, Moser C, Schatz S, Kamolz LP, Magnes C, Sinner F, Sedej S, Frohlich KU, Juhasz G, Pieber TR, Dengjel J, et al. (2014). Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell metabolism 19(3): 431-444. https://doi.org/10.1016/j.cmet.2014.02.010