
Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.

Luminal acetylation of microtubules is not essential for Plasmodium berghei and Toxoplasma gondii survival
Acetylation of α-tubulin at lysine 40 is not essential for cytoskeletal stability in Plasmodium berghei or Toxoplasma gondii, suggesting redundancy and plasticity in microtubule regulation in these parasites.

The dual-site agonist for human M2 muscarinic receptors Iper-8-naphtalimide induces mitochondrial dysfunction in Saccharomyces cerevisiae
S. cerevisiae is a model to study human GPCRs. N-8-Iper, active against glioblastoma via M2 receptor, causes mitochondrial damage in yeast by binding Ste2, highlighting evolutionary conservation of GPCRs.

Integrative Omics reveals changes in the cellular landscape of peroxisome-deficient pex3 yeast cells
To uncover the consequences of peroxisome deficiency, we compared Saccharomyces cerevisiae wild-type with pex3 cells, which lack peroxisomes, employing quantitative proteomics and transcriptomics technologies.

Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
Rebekkah E. Pope1, Patrick Ballmann2, Lisa Whitworth3 and Rolf A. Prade1,*
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
Evelyn Tevere1,a, María G. Mediavilla1,a, Cecilia B. Di Capua1, Marcelo L. Merli1, Carlos Robello2,3, Luisa Berná2,4 and Julia A. Cricco
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.
Sir2 regulates selective autophagy in stationary-phase yeast cells
Ji-In Ryua, Juhye Junga, and Jeong-Yoon Kim
This study establishes Sir2 as a previously unrecognized regulator of selective autophagy during the stationary phase and highlight how cells dynamically control organelle degradation.
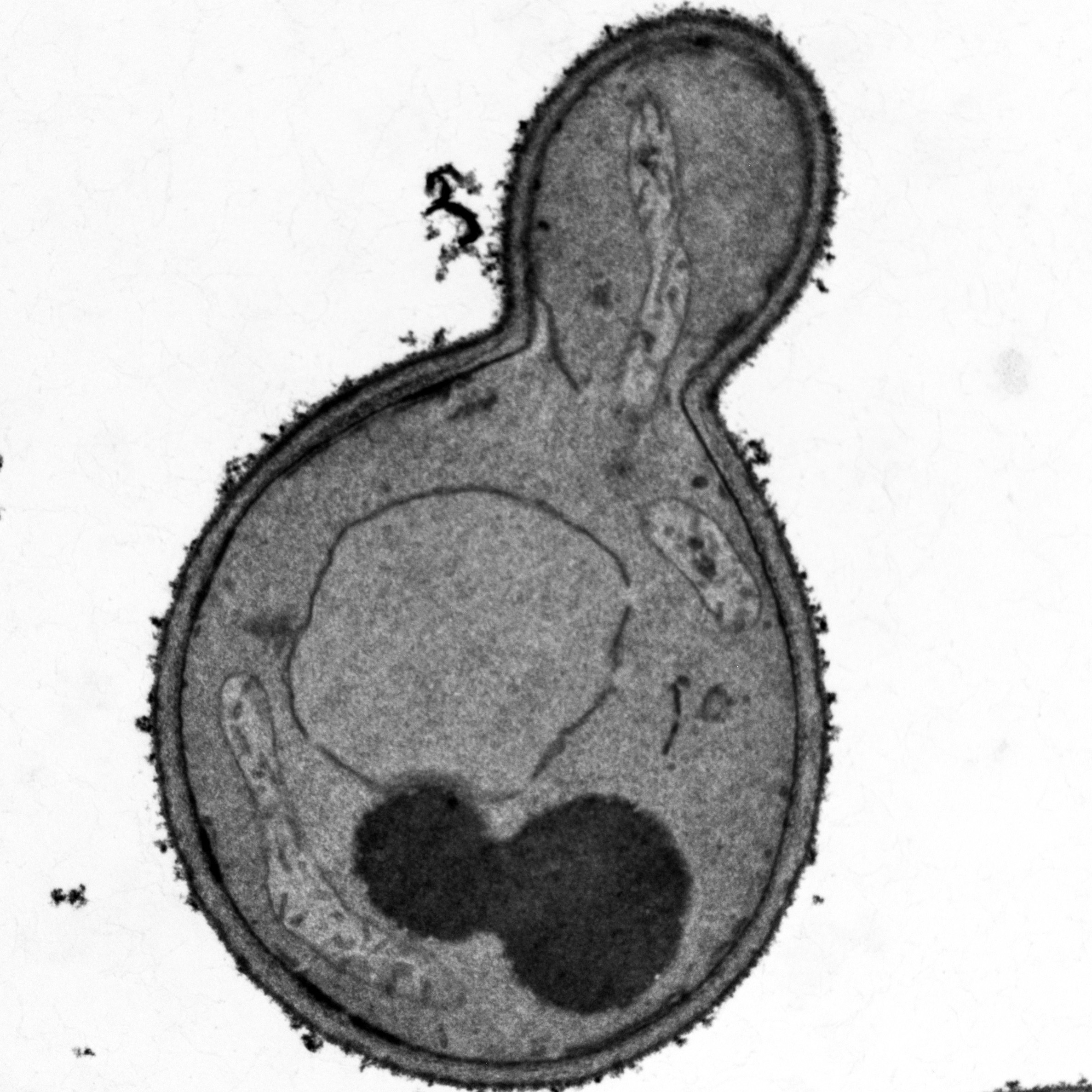
Microwave-assisted preparation of yeast cells for ultrastructural analysis by electron microscopy
Moritz Mayera, Christina Schuga, Stefan Geimer, Till Klecker and Benedikt Westermann
Budding yeast Saccharomyces cerevisiae is widely used as a model organism to study the biogenesis and architecture of organellar membranes, which can be visualized by transmission electron microscopy (TEM).

A complex remodeling of cellular homeostasis distinguishes RSV/SARS-CoV-2 co-infected A549-hACE2 expressing cell lines
Claudia Vanetti1, Irma Saulle1,2, Valentina Artusa1,2, Claudia Moscheni1, Gioia Cappelletti1, Silvia Zecchini1, Sergio Strizzi1, Micaela Garziano1,2, Claudio Fenizia1,2, Antonella Tosoni1, Martina Broggiato1, Pasquale Ogno1, Manuela Nebuloni1, Mario Clerici2,3, Daria Trabattoni1, Fiona Limanaqi1 and Mara Biasin1
Given the common tropism of SARS-CoV-2 and RSV, and the unclear consequences of their mutual influence, we developed an in vitro lung epithelial cell model to study the molecular mechanisms and cellular pathways modulated in viral co-infection.

Fecal gelatinase does not predict mortality in patients with alcohol-associated hepatitis
Yongqiang Yang1,a, Philipp Hartmann2,3,a and Bernd Schnabl1,4
This study aimed to investigate the significance of fecal gelatinase on clinical outcomes in patients with alcohol-associated hepatitis. In conclusion, in our cohort, fecal gelatinase does not predict mortality and does not indicate higher disease severity in patients with alcohol-associated hepatitis.

Direct detection of stringent alarmones (pp)pGpp using malachite green
Muriel Schicketanz1, Magdalena Petrová2, Dominik Rejman2, Margherita Sosio3, Stefano Donadio3 and Yong Everett Zhang1
In this study, we demonstrate the surprising discovery of a commercially available, low-cost malachite green (MG) detection kit, originally designed for orthophosphate (Pi) detection, for detecting (p)ppGpp and its analogues, especially pGpp

Promoter methylation and increased expression of PD-L1 in patients with active tuberculosis
Yen-Han Tseng1,2, Sheng-Wei Pan1,2,3, Jhong-Ru Huang2,4, Chang-Ching Lee1, Jung-Jyh Hung2,5, Po-Kuei Hsu2,5, Nien-Jung Chen6, Wei-Juin Su2,7, Yuh-Min Chen1,2 and Jia-Yih Feng1,2,8
The PD-1/PD-L1 pathway plays a pivotal role in T cell activity and is involved in the pathophysiology of tuberculosis. Here we show that PD-L1 expression is increased in patients with active tuberculosis and is correlated with treatment outcomes.

Quantification methods of Candida albicans are independent irrespective of fungal morphology
Amanda B Soares1, Maria C de Albuquerque1, Leticia M Rosa1, Marlise I Klein 2, Ana C Paravina1, Paula A Barbugli1, Livia N Dovigo3 and Ewerton G de O Mima1
Our study demonstrated that the quantification methods of C. albicans (cells/mL, CFU/mL, and vPCR) did not agree, regardless of the fungal morphology/growth, even though a significant and strong correlation is observed.

Pathogenic Escherichia coli change the adhesion between neutrophils and endotheliocytes in the experimental bacteremia model
Svetlana N Pleskova1,2,*, Nikolay A Bezrukov1, Sergey Z Bobyk1, Ekaterina N Gorshkova1 and Dimitri V Novikov3
In this work, we have demonstrated that in the model of experimental septicemia there is a disruption of adhesion contacts between neutrophils and endothelial cells, manifested by a decrease in adhesion force and work upon exposure to E. coli.

Arsenite treatment induces Hsp90 aggregates distinct from conventional stress granules in fission yeast
Naofumi Tomimotoa, Teruaki Takasakia and Reiko Sugiura
Given the conserved role of Hsp90 as a molecular chaperone protein, our findings presented in this study may suggest a novel type of arsenite-induced biological condensates, wherein Hsp90 plays a key role in maintaining its integrity.
New insights in the mode of action of anti-leishmanial drugs by using chemical mutagenesis screens coupled to next-generation sequencing
Arijit Bhattacharya1, Sophia Bigot2, Prasad Kottayil Padmanabhan2, Angana Mukherjee2, Adriano Coelho3, Philippe Leprohon2, Barbara Papadopoulou2 and Marc Ouellette2
In this article, the authors comment on the study “Coupling chemical mutagenesis to next generation sequencing for the identification of drug resistance mutations in Leishmania” by Bhattacharya et al. (Nat Commun, 2019), which introduces Mut-seq, a chemical mutagenesis and sequencing approach, to uncover drug resistance mechanisms in Leishmania, revealing links between lipid metabolism genes and miltefosine resistance, and a protein kinase involved in translation conferring paromomycin resistance.
Microfluidic techniques for separation of bacterial cells via taxis
Jyoti P. Gurung1, Murat Gel2,3 and Matthew A. B. Baker1,3
Microfluidic tools, ideal for studying microbial motility due to their control over laminar flows at microscopic scales, enable precise analysis of various taxis behaviors and have advanced applications in synthetic biology, directed evolution, and medical microbiology.
Influence of delivery and feeding mode in oral fungi colonization – a systematic review
Maria Joao Azevedo1,2,3,4, Maria de Lurdes Pereira1,5, Ricardo Araujo2,3,6, Carla Ramalho3,7,8, Egija Zaura4 and Benedita Sampaio-Maia1,2,3
A systematic review of oral fungal colonization in infants found that while breastfeeding did not significantly affect the oral mycobiome, vaginal delivery was associated with higher oral yeast colonization, particularly of Candida albicans.
A holobiont view on thrombosis: unravelling the microbiota’s influence on arterial thrombus growth
Giulia Pontarollo1, Klytaimnistra Kiouptsi1 and Christoph Reinhardt1,2
In this article, the authors comment on the study “The microbiota promotes arterial thrombosis in low-density lipoprotein receptor-deficient mice” by Kiouptsi et al. (mBio, 2019) that showed that commensal microbiota, intricately linked to host physiology, may influence cardiovascular disease, as shown by studies using germ-free atherosclerosis-prone mice to examine how microbial presence and diet affect arterial thrombosis and lesion development.
The role of Lactobacillus species in the control of Candida via biotrophic interactions
Isabella Zangl1, Ildiko-Julia Pap2, Christoph Aspöck2 and Christoph Schüller1,3
Microbial communities, including Candida and Lactobacillus species, play a crucial role in human health, particularly in the context of mucosal infections, but our understanding of their interactions and effects is still incomplete due to the variability of species and isolates as well as the complexity of the human host.
Tribal warfare: Commensal Neisseria kill pathogen Neisseria gonorrhoeae using its DNA
Magdalene So1 and Maria A. Rendón1
This article comments on work published by Kim et al (Cell Host Microbe, 2019), which adds a new dimension to the concept of commensal protection. It shows that commensal Neisseria kill the closely related pathogen N. gonorrhoeae through an unexpected mechanism, one that involves genetic competence, DNA methylation state and recombination.
Yet another job for the bacterial ribosome
Andrea Origi1,2, Ana Natriashivili1,2, Lara Knüpffer1, Clara Fehrenbach1, Kärt Denks1,2, Rosella Asti1 and Hans-Georg Koch1
This article comments on work published by Knüpffer et al (mBio, 2019), which revealed the intricate interaction of uL23 with yet another essential player in bacteria, the ATPase SecA, which is best known for its role during post-translational secretion of proteins across the bacterial SecYEG translocon
Gut microbial metabolites in depression: understanding the biochemical mechanisms
Giorgia Caspani1, Sidney Kennedy2-5, Jane A. Foster6 and Jonathan Swann1
This article shows how the gut microbiota contributes to the pathophysiology of depression and examines the mechanisms by which microbially-derived molecules may influence depressive behavior, highlighting the potential of dietary interventions as novel therapeutic strategies.
The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg
Joana N. Bugalhão1 and Luís Jaime Mota1
CThis article shows an in-depth review on the current knowledge and outstanding questions about secreted proteins from Chlamydia trachomatis, detailing their roles in host cell interaction and immune response evasion.
The emerging role of complex modifications of tRNALysUUU in signaling pathways
Patrick C. Thiaville1,2,3,4 and Valérie de Crécy-Lagard2,4
This comment discusses the article “Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling” by Scheidt et al (Microbial Cell, 2014).
Only functional localization is faithful localization
Roland Lill1,2,3
This article comments on work published by Peleh et al. (Microbial Cell 2014), which analyzes the localization of Dre2 in Saccharomyces cerevisiae.
One cell, one love: a journal for microbial research
Didac Carmona-Gutierrez1, Guido Kroemer2-6 and Frank Madeo1
In this inaugural article of Microbial Cell, we highlight the importance of microbial research in general and the journal’s intention to serve as a publishing forum that supports and enfolds the scientific diversity in this area as it provides a unique, high-quality and universally accessible source of information and inspiration.
What’s the role of autophagy in trypanosomes?
Katherine Figarella1 and Néstor L. Uzcátegui1,2
This article comments on Proto et al. (Microbial Cell, 2014), who report first insights into the molecular mechanism of autophagy in African trypanosomes by generating reporter bloodstream form cell lines.
Microbial Cell
is an open-access, peer-reviewed journal that publishes exceptionally relevant research works that implement the use of unicellular organisms (and multicellular microorganisms) to understand cellular responses to internal and external stimuli and/or human diseases.
you can trust
Can’t find what you’re looking for?
You can browse all our issues and published articles here.
FAQs
Peer-reviewed, open-access research using unicellular organisms (and multicellular microorganisms) to understand cellular responses and human disease.
The journal (founded in 2014) is led by its Editors-in-Chief Frank Madeo, Didac Carmona-Gutierrez, and Guido Kroemer
Microbial Cell has been publishing original scientific literature since 2014, and from the very beginning has been managed by active scientists through an independent Publishing House (Shared science Publishers). The journal was conceived as a platform to acknowledge the importance of unicellular organisms, both as model systems as well as in the biological context of human health and disease.
Ever since, Microbial Cell has very positively developed and strongly grown into a respected journal in the unicellular research community and even beyond. This scientific impact is reflected in the yearly number of citations obtained by articles published in Microbial Cell, as recorded by the Web of Science (Clarivate, formerly Thomson/Reuters):

The scientific impact of Microbial Cell is also mirrored in a series of milestones:
2015: Microbial Cell is included in the Emerging Sources Citation Index (ESCI), a selection of developing journals drafted by Clarivate Analytics based on the candidate’s publishing standards, quality, editorial content, and citation data. Note: As an ESCI-selected journal, Microbial Cell is currently being evaluated in a rigorous and long process to determine an inclusion in the Science Citation Index Expanded (SCIE), which allows the official calculation of Clarivate Analytics’ impact factor.
2016: Microbial Cell is awarded the so-called DOAJ Seal by the selective Directory of Open Access Journals (DOAJ). The DOAJ Seal is an exclusive mark of certification for open access journals granted by DOAJ to journals that adhere to outstanding best practice and achieve an extra high and clear commitment to open access and high publishing standards.
2017: Microbial Cell is included in Pubmed Central (PMC), allowing the archiving of all the journal’s articles in PMC and PubMed.
2019: Microbial Cell is indexed in the prestigious abstract and citation database Scopus after a thorough selection process. This also means that Microbial Cell obtains, for the first time, an official Scopus CiteScore as well as an official journal ranking in the Scimago Journal and Country Ranking.
2022: Microbial Cell’s CiteScore reaches a value of 7.2 for the year 2021, positioning Microbial Cell among the top microbiology journals (previously available CiteScores: 2019: 5.4; 2020: 5.1).
2022: Microbial Cell is indexed in the highly selective Science Citation Index Expanded™, which covers approx. 9,500 of the world’s most impactful journals across 178 scientific disciplines. In their journal selection and curation process, Clarivate´s editors apply 24 ‘quality’ criteria and four ‘impact’ criteria to select the most influential journals in their respective fields. This selection is also a pre-requisite for inclusion in the JCR, which features the impact factor.
2022: Microbial Cell is listed in the Journal Citation Reports™ (JCR), and obtains its first official Journal Impact Factor™ (JIF) for the year 2021: 5.316.Check Article Types and Manuscript Preparation guidelines. Submit online via Scholastica.


Metabolic pathways further increase the complexity of cell size control in budding yeast
Jorrit M. Enserink
This article comments on work published by Soma et al. (Microbial Cell, 2014), which teased apart the effect of metabolism and growth rate on setting of critical cell size in Saccharomyces cerevisiae.