
Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.

Luminal acetylation of microtubules is not essential for Plasmodium berghei and Toxoplasma gondii survival
Acetylation of α-tubulin at lysine 40 is not essential for cytoskeletal stability in Plasmodium berghei or Toxoplasma gondii, suggesting redundancy and plasticity in microtubule regulation in these parasites.

The dual-site agonist for human M2 muscarinic receptors Iper-8-naphtalimide induces mitochondrial dysfunction in Saccharomyces cerevisiae
S. cerevisiae is a model to study human GPCRs. N-8-Iper, active against glioblastoma via M2 receptor, causes mitochondrial damage in yeast by binding Ste2, highlighting evolutionary conservation of GPCRs.

Integrative Omics reveals changes in the cellular landscape of peroxisome-deficient pex3 yeast cells
To uncover the consequences of peroxisome deficiency, we compared Saccharomyces cerevisiae wild-type with pex3 cells, which lack peroxisomes, employing quantitative proteomics and transcriptomics technologies.

Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
Rebekkah E. Pope1, Patrick Ballmann2, Lisa Whitworth3 and Rolf A. Prade1,*
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
Evelyn Tevere1,a, María G. Mediavilla1,a, Cecilia B. Di Capua1, Marcelo L. Merli1, Carlos Robello2,3, Luisa Berná2,4 and Julia A. Cricco
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.
Sir2 regulates selective autophagy in stationary-phase yeast cells
Ji-In Ryua, Juhye Junga, and Jeong-Yoon Kim
This study establishes Sir2 as a previously unrecognized regulator of selective autophagy during the stationary phase and highlight how cells dynamically control organelle degradation.

Unresolved mystery of cyclic nucleotide second messengers, periplasmic acid phosphatases and bacterial natural competence
Kristina Kronborg and Yong Everett Zhang
In this study we aimed to identify the promotors responsible for the expression of the non-specific acid phosphatase AphA during different starvation conditions, to confirm the requirement of the cAMP-dependent CRP regulon for aphA expression, and to finally identify regulators of its expression.

Polyadenylated versions of small non-coding RNAs in Saccharomyces cerevisiae are degraded by Rrp6p/Rrp47p independent of the core nuclear exosome
Anusha Chaudhuri1,#, Soumita Paul2,#, Mayukh Banerjea2 and Biswadip Das2
In this investigation, we unveiled a novel functional role of the major nuclear 3′→5′ exoribonuclease, Rrp6p, and its cofactor Rrp47p in the degradation of polyadenylated versions of several mature sncRNAs, including 5S, 5.8S rRNAs, all sn- and some select snoRNAs in the baker’s yeast S. cerevisiae.
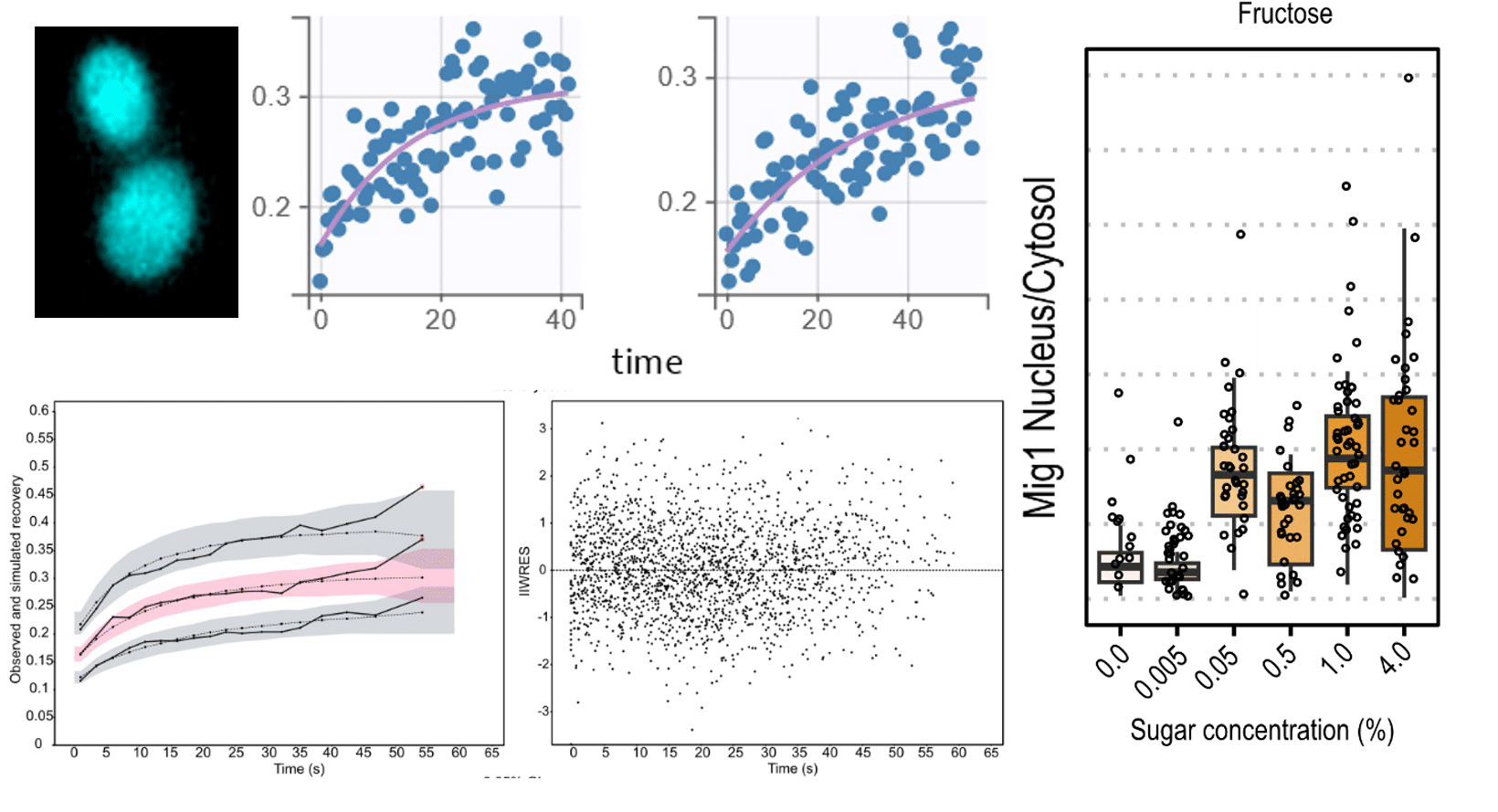
Exploring carbon source related localization and phosphorylation in the Snf1/Mig1 network using population and single cell-based approaches
Svenja Braam1, Farida Tripodi2, Linnea Österberg1,3, Sebastian Persson1, Niek Welkenhuysen1, Paola Coccetti2 and Marija Cvijovic1
In this work we set out to explore the relationship between the subcellular localization and regulation of kinases in the context of carbon source signaling. The data presented in this paper reinforce the notion that not only the activation/inactivation of kinases but also their subcellular localization and that of their targets influence fate decisions in response to environmental changes.

A Modular Cloning Toolkit for the production of recombinant proteins in Leishmania tarentolae
Katrin Hieronimus1,2,#, Tabea Donauer1,2,#, Jonas Klein1,#, Bastian Hinkel1,#, Julia Vanessa Spänle1,#, Anna Probst1,#, Justus Niemeyer1,#, Salina Kibrom1, Anna Maria Kiefer1, Luzia Schneider2, Britta Husemann2, Eileen Bischoff2, Sophie Möhring2, Nicolas Bayer1, Dorothée Klein1, Adrian Engels1, Benjamin Gustav Ziehmer2, Julian Stieß3, Pavlo Moroka1, Michael Schroda1, and Marcel Deponte2
Modular Cloning (MoClo) is based on libraries of standardized genetic parts that can be directionally assembled via Golden Gate cloning in one-pot reactions into transcription units and multigene constructs. We established a MoClo toolkit and exemplified its application for the production of recombinant proteins in L. tarentolae.
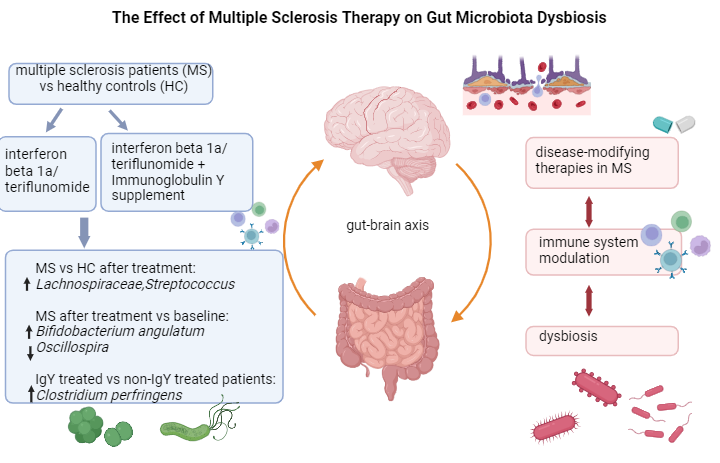
The effect of multiple sclerosis therapy on gut microbiota dysbiosis: a longitudinal prospective study
Andreea-Cristina Paraschiv1,a, Vitalie Vacaras1,2,a, Cristina Nistor1,2, Cristiana Vacaras3, Stefan Strilciuc1 and Dafin F Muresanu1,2
The gut microbiota, a complex ecosystem with various immune functions, plays a significant role in MS, and its response to different treatments is highlighted in this study. In clinical practice, maintaining a healthy microbiota is crucial for individuals with MS.
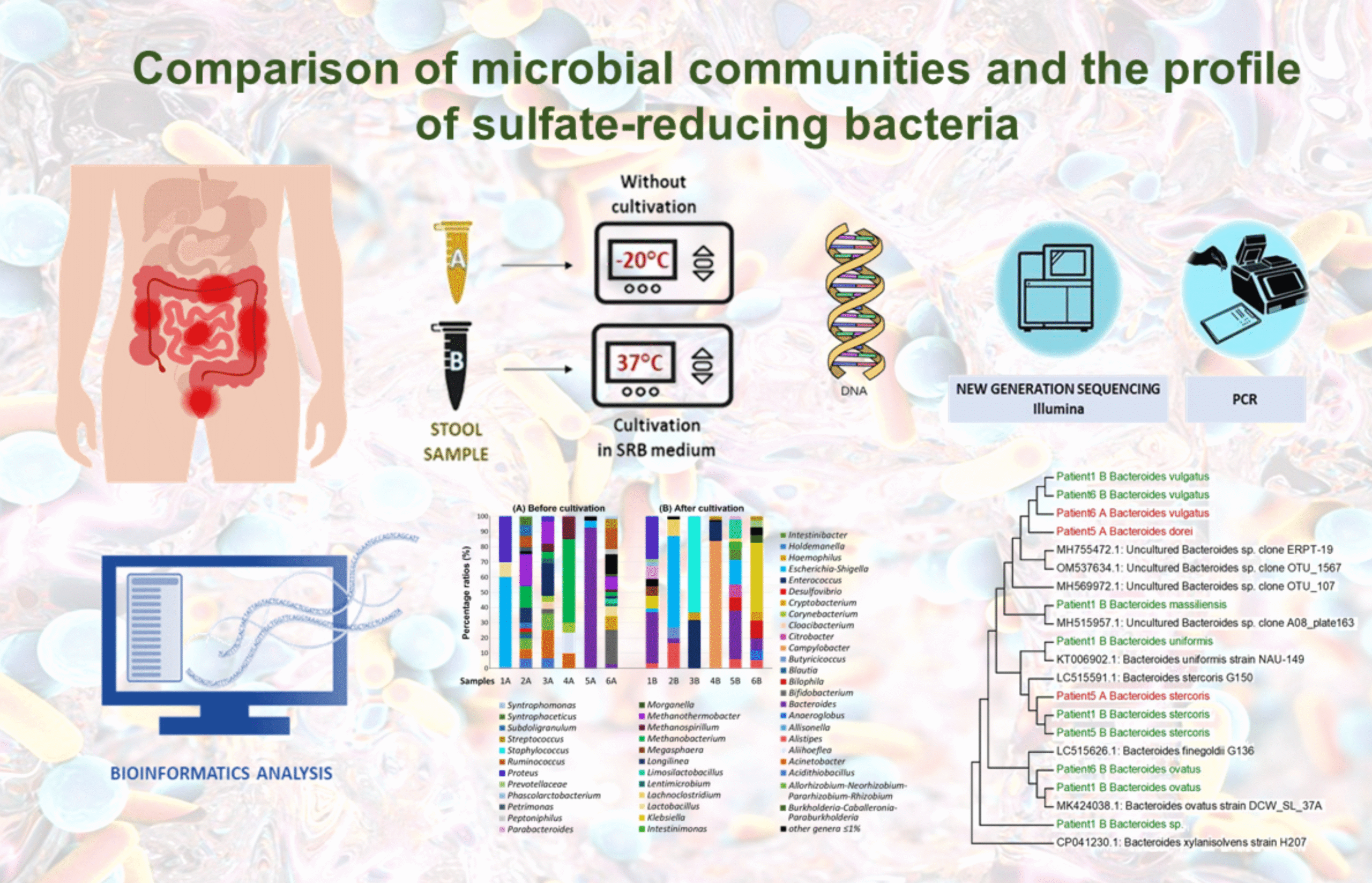
Comparison of microbial communities and the profile of sulfate-reducing bacteria in patients with ulcerative colitis and their association with bowel diseases: a pilot study
Ivan Kushkevych1, Kristýna Martínková1, Lenka Mráková1, Francesco Giudici2, Simone Baldi2, David Novak3, Márió Gajdács4, Monika Vítězová1, Dani Dordevic5, Amedeo Amedei2 and Simon K.-M. R. Rittmann6
Considerable evidence has accumulated regarding the molecular relationship between gut microbiota (GM) composition and the onset (clinical presentation and prognosis) of ulcerative colitis UC. Our findings highlight, among other observations, significant variations in the gut microbial composition among patients with varying disease severity and activity.

Replicative aging in yeast involves dynamic intron retention patterns associated with mRNA processing/export and protein ubiquitination
Jesús Gómez-Montalvo1, Alvaro de Obeso Fernández del Valle1, Luis Fernando De la Cruz Gutiérrez1, Jose Mario Gonzalez-Meljem1 and Christian Quintus Scheckhuber1
Saccharomyces cerevisiae has yielded relevant insights into some of the basic mechanisms of organismal aging. Among these are genomic instability, oxidative stress, caloric restriction and mitochondrial dysfunction. Our work uncovers a previously unexplored layer of the transcriptional program of yeast aging and, more generally, expands the knowledge on the occurrence of alternative splicing in baker´s yeast.
Transcriptional and genomic mayhem due to aging-induced nucleosome loss in budding yeast
Zheng Hu1, Kaifu Chen2, Wei Li2 and Jessica K. Tyler2
This article comments on work published by Zheng et al. (Genes and Development, 2014), which investigated a loss of histones during replicative aging in budding yeast, which was also accompanied by a significantly-increased frequency of genomic instability including DNA breaks, chromosomal translocations, retrotransposition, and transfer of mitochondrial DNA into the nuclear genome.
The Parkinson’s disease-associated protein α-synuclein disrupts stress signaling – a possible implication for methamphetamine use?
Shaoxiao Wang1 and Stephan N. Witt1,2
This article comments on work published by Wang et al. (PNAS, 2012), which reported that human α-syn, at high expression levels, disrupts stress-activated signal transduction pathways in both yeast and human neuroblastoma cells. Disruption of these signaling pathways ultimately leads to vulnerability to stress and to cell death.
Massive gene swamping among cheese-making Penicillium fungi
Jeanne Ropars1,2, Gabriela Aguileta1,2,3, Damien M. de Vienne4,5 and Tatiana Giraud1,2
This article comments on work published by Cheeseman et al. (Nat Comm, 2014), which indicates that horizontal gene transfer is a crucial mechanism of rapid adaptation, even among eukaryotes.
Genome-wide studies of telomere biology in budding yeast
Yaniv Harari and Martin Kupiec
In the last decade, technical advances have allowed carrying out systematic genome-wide screens for mutants affecting various aspects of telomere biology. In this review we summarize these efforts, and the insights that this Systems Biology approach has produced so far.
Mnemons: encoding memory by protein super-assembly
Fabrice Caudron and Yves Barral
This article comments on work published by Caudron and Barral (Cell, 2013), which proposes that polyQ- and polyN-based elements, termed mnemons, act as cellular memory devices to encode previous environmental conditions.
Intersubunit communications within KaiC hexamers contribute the robust rhythmicity of the cyanobacterial circadian clock
Yohko Kitayama1, Taeko Nishiwaki-Ohkawa1,2 and Takao Kondo1
This article comments on work published by Kitayama et al. (Nat Comm, 2013), which suggests that intersubunit communication precisely synchronizes KaiC subunits to avoid dephasing, and contributes to the robustness of circadian rhythms in cyanobacteria.
Mitochondrial protein import under kinase surveillance
Magdalena Opalińska1 and Chris Meisinger1,2
This article summarizes recent discoveries in the yeast Saccharomyces cerevisiae model system that point towards a vital role of reversible phosphorylation in regulation of mitochondrial protein import.
Building a flagellum in biological outer space
Lewis D. B. Evans, Colin Hughes and Gillian M. Fraser
This article comments on work published by Evans et al. (Nature, 2013), which presents a simple and elegant transit mechanism in which growth is powered by the subunits themselves as they link head-to-tail in a chain that is pulled through the length of the growing structure to the tip. This new mechanism answers an old question and may have resonance in other assembly processes.
Transceptors as a functional link of transporters and receptors
George Diallinas
A relative newcomer in environment sensing are the so called transceptors, membrane proteins that possess both solute transport and receptor-like signaling activities. Now, the transceptor concept is further enlarged to include micronutrient sensing via the iron and zinc high-affinity transporters of Saccharomyces cerevisiae.
S. pombe placed on the prion map
Jacqueline Hayles
This article comments on work published by Sideri et al. (Microbial Cell, 2017), which identified the Ctr4 prion in S. pombe.
Using microbes as a key tool to unravel the mechanism of autophagy and the functions of the ATG proteins
Mario Mauthe1,2 and Fulvio Reggiori1,2
Microbes have served to discover and characterize unconventional functions of the ATG proteins, which are uncoupled from their role in autophagy. In our recent study, we have taken advantage of viruses as a screening tool to determine the extent of the unconventional functions of the ATG proteome and characterize one of them.
Autophagy: one more Nobel Prize for yeast
Andreas Zimmermann1, Katharina Kainz1, Aleksandra Andryushkova1, Sebastian Hofer1, Frank Madeo1,2 and Didac Carmona-Gutierrez1
The recent announcement of the 2016 Nobel Prize in Physiology or Medicine, awarded to Yoshinori Ohsumifor the discoveries of mechanisms governing autophagy, underscores the importance of intracellular degradation and recycling. Here we provide a quick historical overview that mirrors both the importance of autophagy as a conserved and essential process for cellular life and death as well as the crucial role of yeast in its mechanistic characterization.
Physiology, phylogeny, and LUCA
William F. Martin1,2, Madeline C. Weiss1, Sinje Neukirchen3, Shijulal Nelson-Sathi4, Filipa L. Sousa3
Genomes record their own history. But if we want to look all the way back to life’s beginnings some 4 billion years ago, the record of microbial evolution that is preserved in prokaryotic genomes is not easy to read. The classical approach has been to look for genes that are universally distributed. Another approach is to make all trees for all genes, and sift out the trees where signals have been overwritten by lateral gene transfer. What is left ought to be ancient. If we do that, what do we find?
Sexually transmitted infections: old foes on the rise
Didac Carmona-Gutierrez1,*, Katharina Kainz1 and Frank Madeo1,2,*
Sexually transmitted infections (STIs) are commonly spread via sexual contact. It is estimated that one million STIs are acquired every day worldwide. Besides their impact on sexual, reproductive and neonatal health, they can cause disastrous and life-threatening complications if left untreated. In addition to this personal burden, STIs also represent a socioeconomic problem, deriving in treatment costs of tremendous proportions. Despite a substantial progress in diagnosis, treatment and prevention, the incidence of many common STIs is increasing, and STIs continue to represent a global public health problem and a major cause for morbidity and mortality. With this Special Issue, Microbial Cell provides an in-depth overview of the eight major STIs, covering all relevant features of each infection.
Microbial Cell
is an open-access, peer-reviewed journal that publishes exceptionally relevant research works that implement the use of unicellular organisms (and multicellular microorganisms) to understand cellular responses to internal and external stimuli and/or human diseases.
you can trust
Can’t find what you’re looking for?
You can browse all our issues and published articles here.
FAQs
Peer-reviewed, open-access research using unicellular organisms (and multicellular microorganisms) to understand cellular responses and human disease.
The journal (founded in 2014) is led by its Editors-in-Chief Frank Madeo, Didac Carmona-Gutierrez, and Guido Kroemer
Microbial Cell has been publishing original scientific literature since 2014, and from the very beginning has been managed by active scientists through an independent Publishing House (Shared science Publishers). The journal was conceived as a platform to acknowledge the importance of unicellular organisms, both as model systems as well as in the biological context of human health and disease.
Ever since, Microbial Cell has very positively developed and strongly grown into a respected journal in the unicellular research community and even beyond. This scientific impact is reflected in the yearly number of citations obtained by articles published in Microbial Cell, as recorded by the Web of Science (Clarivate, formerly Thomson/Reuters):

The scientific impact of Microbial Cell is also mirrored in a series of milestones:
2015: Microbial Cell is included in the Emerging Sources Citation Index (ESCI), a selection of developing journals drafted by Clarivate Analytics based on the candidate’s publishing standards, quality, editorial content, and citation data. Note: As an ESCI-selected journal, Microbial Cell is currently being evaluated in a rigorous and long process to determine an inclusion in the Science Citation Index Expanded (SCIE), which allows the official calculation of Clarivate Analytics’ impact factor.
2016: Microbial Cell is awarded the so-called DOAJ Seal by the selective Directory of Open Access Journals (DOAJ). The DOAJ Seal is an exclusive mark of certification for open access journals granted by DOAJ to journals that adhere to outstanding best practice and achieve an extra high and clear commitment to open access and high publishing standards.
2017: Microbial Cell is included in Pubmed Central (PMC), allowing the archiving of all the journal’s articles in PMC and PubMed.
2019: Microbial Cell is indexed in the prestigious abstract and citation database Scopus after a thorough selection process. This also means that Microbial Cell obtains, for the first time, an official Scopus CiteScore as well as an official journal ranking in the Scimago Journal and Country Ranking.
2022: Microbial Cell’s CiteScore reaches a value of 7.2 for the year 2021, positioning Microbial Cell among the top microbiology journals (previously available CiteScores: 2019: 5.4; 2020: 5.1).
2022: Microbial Cell is indexed in the highly selective Science Citation Index Expanded™, which covers approx. 9,500 of the world’s most impactful journals across 178 scientific disciplines. In their journal selection and curation process, Clarivate´s editors apply 24 ‘quality’ criteria and four ‘impact’ criteria to select the most influential journals in their respective fields. This selection is also a pre-requisite for inclusion in the JCR, which features the impact factor.
2022: Microbial Cell is listed in the Journal Citation Reports™ (JCR), and obtains its first official Journal Impact Factor™ (JIF) for the year 2021: 5.316.Check Article Types and Manuscript Preparation guidelines. Submit online via Scholastica.



Staphylococcus aureus type I signal peptidase: essential or not essential, that’s the question
Wouter L.W. Hazenbos1, Elizabeth Skippington2 and Man-Wah Tan1
This article comments on work published by Morisaki et al. (mBio, 2016), which characterized a novel ABC transporter. This transporter apparently compensates for SpsB’s essential function by mediating alternative cleavage of a subset of proteins at a site distinct from the SpsB-cleavage site, leading to SpsB-independent secretion.