
Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.

Luminal acetylation of microtubules is not essential for Plasmodium berghei and Toxoplasma gondii survival
Acetylation of α-tubulin at lysine 40 is not essential for cytoskeletal stability in Plasmodium berghei or Toxoplasma gondii, suggesting redundancy and plasticity in microtubule regulation in these parasites.

The dual-site agonist for human M2 muscarinic receptors Iper-8-naphtalimide induces mitochondrial dysfunction in Saccharomyces cerevisiae
S. cerevisiae is a model to study human GPCRs. N-8-Iper, active against glioblastoma via M2 receptor, causes mitochondrial damage in yeast by binding Ste2, highlighting evolutionary conservation of GPCRs.

Integrative Omics reveals changes in the cellular landscape of peroxisome-deficient pex3 yeast cells
To uncover the consequences of peroxisome deficiency, we compared Saccharomyces cerevisiae wild-type with pex3 cells, which lack peroxisomes, employing quantitative proteomics and transcriptomics technologies.

Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
Rebekkah E. Pope1, Patrick Ballmann2, Lisa Whitworth3 and Rolf A. Prade1,*
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
Evelyn Tevere1,a, María G. Mediavilla1,a, Cecilia B. Di Capua1, Marcelo L. Merli1, Carlos Robello2,3, Luisa Berná2,4 and Julia A. Cricco
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.
Sir2 regulates selective autophagy in stationary-phase yeast cells
Ji-In Ryua, Juhye Junga, and Jeong-Yoon Kim
This study establishes Sir2 as a previously unrecognized regulator of selective autophagy during the stationary phase and highlight how cells dynamically control organelle degradation.
Chromosome-condensed G1 phase yeast cells are tolerant to desiccation stress
Zhaojie Zhang1 and Gracie R. Zhang2
The budding yeast Saccharomyces cerevisiae is capable of surviving extreme water loss for a long time. However, less is known about the mechanism of its desiccation tolerance. In this study, we revealed that in an exponential culture, all desiccation tolerant yeast cells were in G1 phase and had condensed chromosomes. (…)
Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its first variants in fourplex real-time quantitative reverse transcription-PCR assays
Mathieu Durand1, Philippe Thibault1, Simon Lévesque2,3, Ariane Brault4, Alex Carignan2, Louis Valiquette2, Philippe Martin2 and Simon Labbé4
The early diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections is required to identify and isolate contagious patients to prevent further transmission of SARS-CoV-2. In this study, we present a multitarget real-time TaqMan reverse transcription PCR (rRT-PCR) assay for the quantitative detection of SARS-CoV-2 and some of its circulating variants harboring mutations that give the virus a selective advantage. Seven different primer-probe sets that included probes containing locked nucleic acid (LNA) nucleotides were designed to amplify specific wild-type and mutant sequences in Orf1ab, Envelope (E), Spike (S), and Nucleocapsid (N) genes (…)
Forced association of SARS-CoV-2 proteins with the yeast proteome perturb vesicle trafficking
Cinzia Klemm1,#, Henry Wood1,#, Grace Heredge Thomas1,#, Guðjón Ólafsson1,2, Mara Teixeira Torres1 and Peter H. Thorpe1
This work demonstrates that the yeast Synthetic Physical Interactions method is a rapid way to identify potential functions of ectopic viral proteins.
Airborne bacteria in show caves from Southern Spain
Irene Dominguez-Moñino1, Valme Jurado1, Miguel Angel Rogerio-Candelera1, Bernardo Hermosin1 and Cesareo Saiz-Jimenez1
This study analyzes the factors conditioning the diversity of airborne bacteria recorded in three Andalusian show caves, subjected to different managements.
Landscapes and bacterial signatures of mucosa-associated intestinal microbiota in Chilean and Spanish patients with inflammatory bowel disease
Nayaret Chamorro1,#, David A. Montero1,2,#, Pablo Gallardo3, Mauricio Farfán3, Mauricio Contreras4, Marjorie De la Fuente2, Karen Dubois2, Marcela A. Hermoso2, Rodrigo Quera5,6, Marjorie Pizarro-Guajardo7,8,9, Daniel Paredes-Sabja7,8,9, Daniel Ginard10, Ramon Rosselló-Móra11 and Roberto Vidal1,8,12
This study investigates the landscapes and alterations of mucosa-associated intestinal microbiota in patients with inflammatory bowel diseases, which cause chronic inflammation of the gut, including ulcerative colitis and Crohn’s disease.
Genome, transcriptome and secretome analyses of the antagonistic, yeast-like fungus Aureobasidium pullulans to identify potential biocontrol genes
Maria Paula Rueda-Mejia1, Lukas Nägeli1, Stefanie Lutz2, Richard D. Hayes3, Adithi R. Varadarajan2, Igor V. Grigoriev3,4, Christian H. Ahrens2,5 and Florian M. Freimoser1
This study highlights the value of a sequential approach starting with genome mining and consecutive transcriptome and secretome analyses in order to identify a limited number of potential target genes for detailed, functional analyses in Aureobasidium pullulans.
Proanthocyanidin-enriched cranberry extract induces resilient bacterial community dynamics in a gnotobiotic mouse model
Catherine C. Neto1,2,#, Benedikt M. Mortzfeld3,#, John R. Turbitt1,2, Shakti K. Bhattarai3, Vladimir Yeliseyev4, Nicholas DiBenedetto4, Lynn Bry4 and Vanni Bucci2,3
This study investigates the effect of a water-soluble, proanthocyanidin-rich cranberry juice extract on the short-term dynamics of a human-derived bacterial community in a gnotobiotic mouse model.
Dry biocleaning of artwork: an innovative methodology for Cultural Heritage recovery?
Giancarlo Ranalli1, Pilar Bosch-Roig2, Simone Crudele1, Laura Rampazzi3,4, Cristina Corti3 and Elisabetta Zanardini5
This work proposes an innovative methodology based on applied biotechnology for the recovery of altered stonework: the “dry biocleaning”, which envisages the use of dehydrated microbial cells without the use of free water or gel-based matrices.
Gut microbiota and ankylosing spondylitis: current insights and future challenges
Andrei Lobiuc1, Liliana Groppa2, Lia Chislari2, Eugeniu Russu2,3, Marinela Homitchi2,3, Camelia Ciorescu2,3, Sevag Hamamah4, I. Codruta Bran1 and Mihai Covasa1
This review explores the growing role of gut microbiota in AS and its potential to reshape targeted treatment strategies and facilitate development of adjunct therapies to address disease onset and progression.
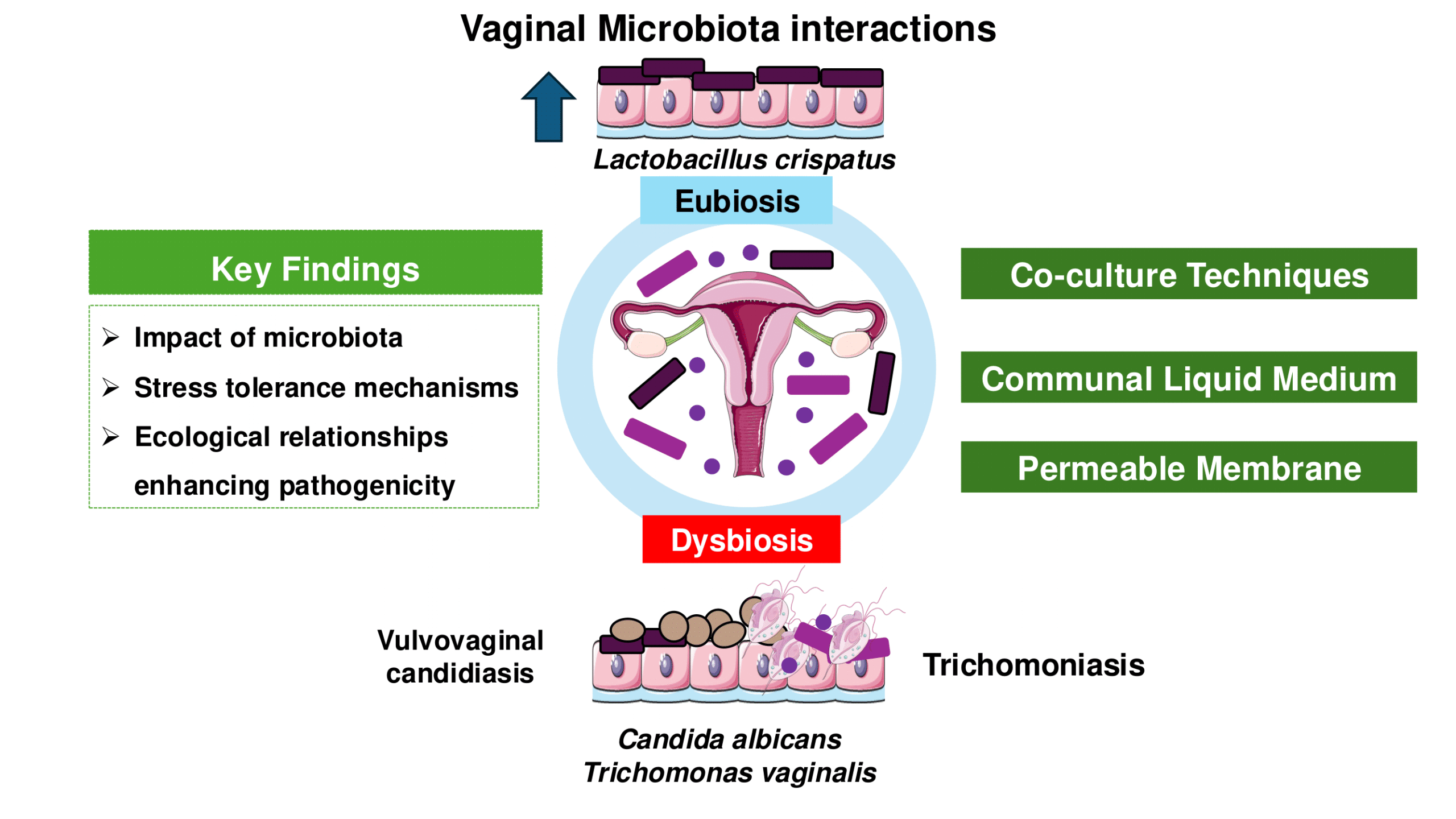
Advancements in vaginal microbiota, Trichomonas vaginalis, and vaginal cell interactions: Insights from co-culture assays
Fernanda Gomes Cardoso and Tiana Tasca
This review updates co-culture and co-incubation techniques for studying interactions of Lactobacillus spp., representing a pre-dominant member of the healthy vaginal microbiota; Candida spp., the most abundant yeast in the vagina, and T. vaginalis, responsible for the most widespread nonviral STI worldwide.
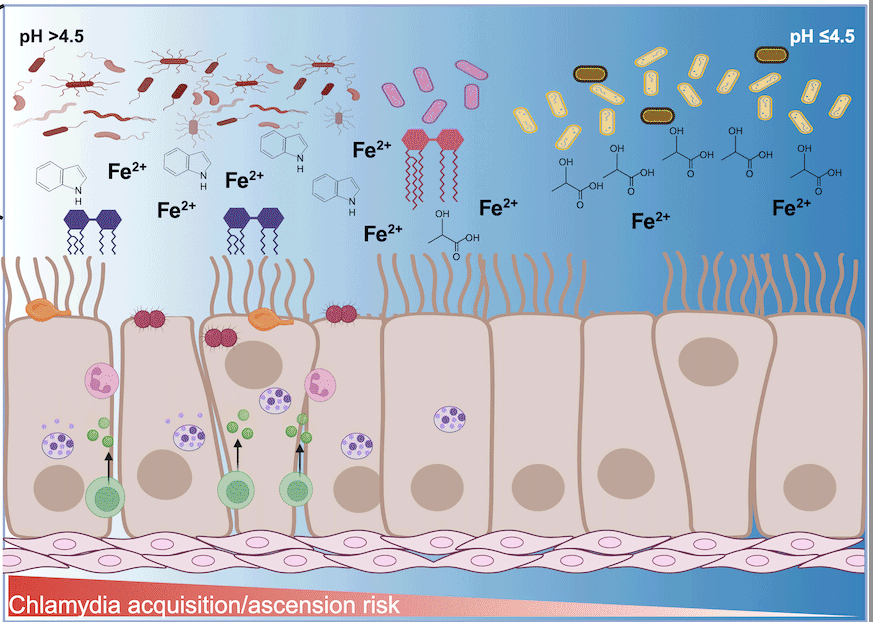
Influence of cervicovaginal microbiota on Chlamydia trachomatis infection dynamics
Emily Hand1, Indriati Hood-Pishchany1,2, Toni Darville1,2 and Catherine M. O’Connell2
This review examines the complex interplay between the cervicovaginal microbiome, C. trachomatis infection, and host immune responses, highlighting the role of metabolites such as short-chain and long-chain fatty acids, indole, and iron in modulating pathogen survival and host defenses.
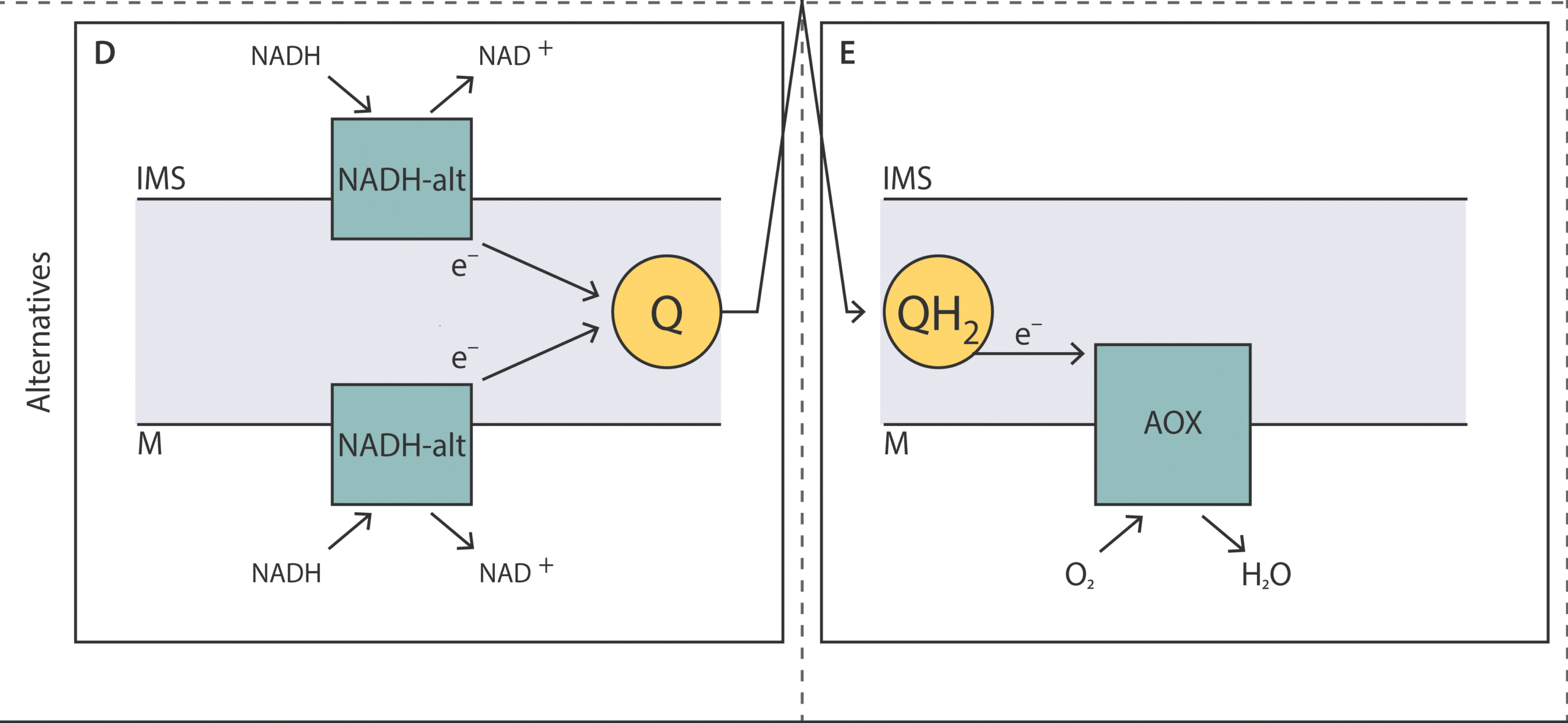
Unveiling the molecular architecture of the mitochondrial respiratory chain of Acanthamoeba castellanii
Christian Q. Scheckhuber1, Sutherland K. Maciver2 and Alvaro de Obeso Fernandez del Valle1
This review provides a comprehensive overview of the mitochondrial res-piratory chain in A. castellanii, focusing on the key alternative components involved in oxidative phosphorylation and their roles in energy metabolism, stress response, and adaptation to various conditions.

Paving the way for new antimicrobial peptides through molecular de-extinction
Karen O. Osiro1, Abel Gil-Ley2, Fabiano C. Fernandes1,3, Kamila B. S. de Oliveira2, Cesar de la Fuente-Nunez4-7, Octavio L. Franco1,2
The advancement of artificial intelligence and molecular de-extinction offers a valuable opportunity not only to discover new antimicrobials but also to provide accurate in silico predictions, thereby shortening the path to addressing the global antibiotic resistance crisis.

Efflux pumps: gatekeepers of antibiotic resistance in Staphylococcus aureus biofilms
Shweta Sinha1, Shifu Aggarwal2,3 and Durg Vijai Singh1
This review aims to elucidate the complex relationship between efflux pumps, antibiotic resistance and biofilm formation in S. aureus with the aim to aid in the development of potential therapeutic targets for combating S. aureus infections, especially those associated with biofilms.

Understanding the molecular mechanisms of human diseases: the benefits of fission yeasts
Lajos Acs-Szabo, Laszlo-Attila Papp and Ida Miklos
Here we collect the latest laboratory protocols and bioinformatics tools for the fission yeasts to highlight the many possibilities available to the research community. In addition, we present several limiting factors that everyone should be aware of when working with yeast models.
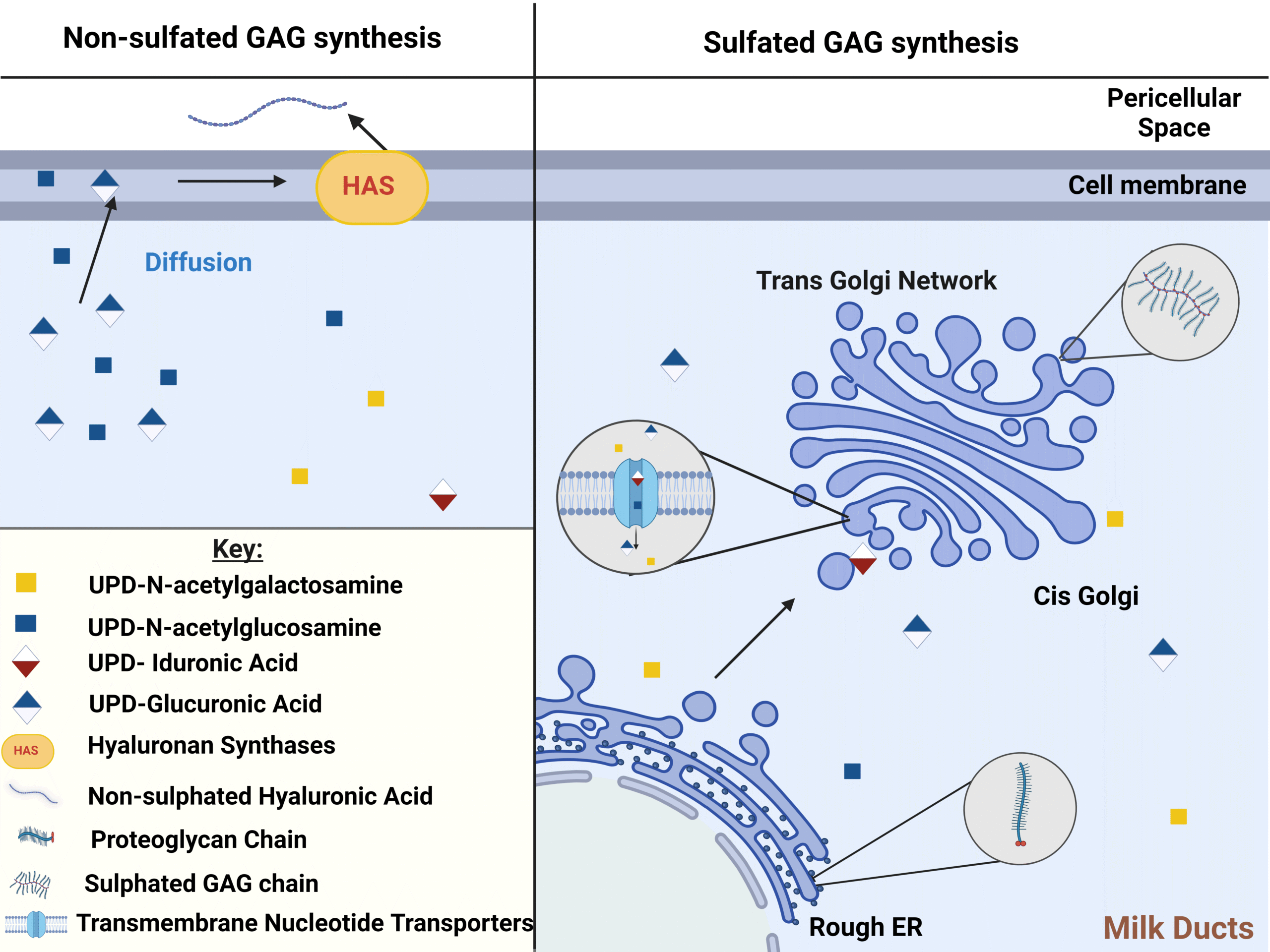
Characterising glycosaminoglycans in human breastmilk and their potential role in infant health
Melissa Greenwood1,2, Patricia Murciano-Martínez3, Janet Berrington4, Sabine L Flitsch5, Sean Austin2 and Christopher Stewart1
Glycosaminoglycans are bioactive components present in breast milk and play a potential key role in determining infant health yet are overlooked by many contemporary studies. This review explores their relevance, use and characterisation techniques.
Non-genetic impact factors on chronological lifespan and stress resistance of baker’s yeast
Michael Sauer and Diethard Mattanovich
This article comments on work published by Bisschops et al. (Microbial Cell, 2015), which illustrates how important the choice of the experimental setup is and how culture conditions influcence cellular aging and survival in biotechnological processes.
What’s old is new again: yeast mutant screens in the era of pooled segregant analysis by genome sequencing
Chris Curtin and Toni Cordente
This article comments on work published by Den Abt et al. (Microbial Cell, 2016), which identified genes involved in ethyl acetate formation in a yeast mutant screen based on a new approach combining repeated rounds of chemical mutagenesis and pooled segregant analysis by whole genome sequencing.
The complexities of bacterial-fungal interactions in the mammalian gastrointestinal tract
Eduardo Lopez-Medina1 and Andrew Y. Koh2
This article comments on work published by Lopez-Medina et al. (PLoS Pathog, 2015) and Fan et al. (Nat Med, 2015), which utilize an “artificial” niche, the antibiotic-treated gut with concomitant pathogenic microbe expansion, to gain insight in bacterial-fungal interactions in clinically common scenarios.
Gearing up for survival – HSP-containing granules accumulate in quiescent cells and promote survival
Ruofan Yu and Weiwei Dang
This article comments on work published by Lee et al. (Microbial Cell, 2016), which reports that distinct granules are formed in quiescent and non-quiescent cells, which determines their respective cell fates.
Yeast screening platform identifies FDA-approved drugs that reduce Aβ oligomerization
Triana Amen1,2 and Daniel Kaganovich1
This article comments on work published by Park et al. (Microbial Cell, 2016), which discovered a number of small molecules capable of modulating Aβ aggregation in a yeast model.
Groupthink: chromosomal clustering during transcriptional memory
Kevin A. Morano
In this article, the authors comment on the study “NO1 transcriptional memory leads to DNA zip code-dependent interchromosomal clustering.” by Brickner et al. (Microbial Cell, 2015), discussing the importance and molecular mechanisms of chromosomal clustering during transcriptional memory.
Yeast proteinopathy models: a robust tool for deciphering the basis of neurodegeneration
Amit Shrestha1, 2 and Lynn A. Megeney1, 2, 3
Protein quality control or proteostasis is an essential determinant of basic cell health and aging. Eukaryotic cells have evolved a number of proteostatic mechanisms to ensure that proteins retain functional conformation, or are rapidly degraded when proteins misfold or self-aggregate. This article discusses the use of budding yeast as a robust proxy to study the intersection between proteostasis and neurodegenerative disease.
Microbial Cell
is an open-access, peer-reviewed journal that publishes exceptionally relevant research works that implement the use of unicellular organisms (and multicellular microorganisms) to understand cellular responses to internal and external stimuli and/or human diseases.
you can trust
Can’t find what you’re looking for?
You can browse all our issues and published articles here.
FAQs
Peer-reviewed, open-access research using unicellular organisms (and multicellular microorganisms) to understand cellular responses and human disease.
The journal (founded in 2014) is led by its Editors-in-Chief Frank Madeo, Didac Carmona-Gutierrez, and Guido Kroemer
Microbial Cell has been publishing original scientific literature since 2014, and from the very beginning has been managed by active scientists through an independent Publishing House (Shared science Publishers). The journal was conceived as a platform to acknowledge the importance of unicellular organisms, both as model systems as well as in the biological context of human health and disease.
Ever since, Microbial Cell has very positively developed and strongly grown into a respected journal in the unicellular research community and even beyond. This scientific impact is reflected in the yearly number of citations obtained by articles published in Microbial Cell, as recorded by the Web of Science (Clarivate, formerly Thomson/Reuters):

The scientific impact of Microbial Cell is also mirrored in a series of milestones:
2015: Microbial Cell is included in the Emerging Sources Citation Index (ESCI), a selection of developing journals drafted by Clarivate Analytics based on the candidate’s publishing standards, quality, editorial content, and citation data. Note: As an ESCI-selected journal, Microbial Cell is currently being evaluated in a rigorous and long process to determine an inclusion in the Science Citation Index Expanded (SCIE), which allows the official calculation of Clarivate Analytics’ impact factor.
2016: Microbial Cell is awarded the so-called DOAJ Seal by the selective Directory of Open Access Journals (DOAJ). The DOAJ Seal is an exclusive mark of certification for open access journals granted by DOAJ to journals that adhere to outstanding best practice and achieve an extra high and clear commitment to open access and high publishing standards.
2017: Microbial Cell is included in Pubmed Central (PMC), allowing the archiving of all the journal’s articles in PMC and PubMed.
2019: Microbial Cell is indexed in the prestigious abstract and citation database Scopus after a thorough selection process. This also means that Microbial Cell obtains, for the first time, an official Scopus CiteScore as well as an official journal ranking in the Scimago Journal and Country Ranking.
2022: Microbial Cell’s CiteScore reaches a value of 7.2 for the year 2021, positioning Microbial Cell among the top microbiology journals (previously available CiteScores: 2019: 5.4; 2020: 5.1).
2022: Microbial Cell is indexed in the highly selective Science Citation Index Expanded™, which covers approx. 9,500 of the world’s most impactful journals across 178 scientific disciplines. In their journal selection and curation process, Clarivate´s editors apply 24 ‘quality’ criteria and four ‘impact’ criteria to select the most influential journals in their respective fields. This selection is also a pre-requisite for inclusion in the JCR, which features the impact factor.
2022: Microbial Cell is listed in the Journal Citation Reports™ (JCR), and obtains its first official Journal Impact Factor™ (JIF) for the year 2021: 5.316.Check Article Types and Manuscript Preparation guidelines. Submit online via Scholastica.


Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation.
Saul M. Honigberg
Diploid budding yeast (Saccharomyces cerevisiae) can adopt one of several alternative differentiation fates in response to nutrient limitation, and each of these fates provides distinct biological functions. When different strain backgrounds are taken into account, these various fates occur in response to similar environmental cues, are regulated by the same signal transduction pathways, and share many of the same master regulators. I propose that the relationships between fate choice, environmental cues and signaling pathways are not Boolean, but involve graded levels of signals, pathway activation and master-regulator activity.