Research Articles:
Microbial Cell, Vol. 2, No. 11, pp. 429 - 444; doi: 10.15698/mic2015.11.238
Oxygen availability strongly affects chronological lifespan and thermotolerance in batch cultures of Saccharomyces cerevisiae
1 Department of Biotechnology, Delft University of Technology, Delft, The Netherlands.
2 Instituto de Ciencias de la Vid y del Vino, CSIC, Universidad de La Rioja, Gobierno de La Rioja, Logroño, Spain.
3 Current address: Division of Systems and Synthetic Biology, Department of Biology and Biological Engineering & The Novo Nordisk Foundation Center for Biosustainability, Chalmers University of Technology, Gothenburg, Sweden.
4 Current address: Quercus Europe S.L., L’Hospitalet de Llobregat, Catalonia, Spain.
# Authors contributed equally to this work.
Keywords: chronological lifespan, thermotolerance, stationary phase, anaerobiosis, energetics, transcriptional response, conditioning.
Abbreviations:
CLS - chronological lifespan,
ExP - exponential growth phase,
PD - post-diauxic phase,
ROS - reactive oxygen species,
SP - stationary-phase.
Received originally: 26/06/2015 Received in revised form: 09/09/2015
Accepted: 13/09/2015
Published: 21/10/2015
Correspondence:
Pascale Daran-Lapujade, Julianalaan 67, 2628 BC; Delft, The Netherlands p.a.s.daran-lapujade@tudelft.nl
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Markus M.M. Bisschops, Tim Vos, Rubén Martínez-Moreno, Pilar de la Torre Cortes, Jack T. Pronk, Pascale Daran-Lapujade (2015). Oxygen availability strongly affects chronological lifespan and thermotolerance in batch cultures of Saccharomyces cerevisiae. Microbial Cell2(11): 429-444.
Abstract
Stationary-phase (SP) batch cultures of Saccharomyces cerevisiae, in which growth has been arrested by carbon-source depletion, are widely applied to study chronological lifespan, quiescence and SP-associated robustness. Based on this type of experiments, typically performed under aerobic conditions, several roles of oxygen in aging have been proposed. However, SP in anaerobic yeast cultures has not been investigated in detail. Here, we use the unique capability of S. cerevisiae to grow in the complete absence of oxygen to directly compare SP in aerobic and anaerobic bioreactor cultures. This comparison revealed strong positive effects of oxygen availability on adenylate energy charge, longevity and thermotolerance during SP. A low thermotolerance of anaerobic batch cultures was already evident during the exponential growth phase and, in contrast to the situation in aerobic cultures, was not substantially increased during transition into SP. A combination of physiological and transcriptome analysis showed that the slow post-diauxic growth phase on ethanol, which precedes SP in aerobic, but not in anaerobic cultures, endowed cells with the time and resources needed for inducing longevity and thermotolerance. When combined with literature data on acquisition of longevity and thermotolerance in retentostat cultures, the present study indicates that the fast transition from glucose excess to SP in anaerobic cultures precludes acquisition of longevity and thermotolerance. Moreover, this study demonstrates the importance of a preceding, calorie-restricted conditioning phase in the acquisition of longevity and stress tolerance in SP yeast cultures, irrespective of oxygen availability.
INTRODUCTION
Just like other living organisms, Saccharomyces cerevisiae cells age and have a finite chronological lifespan. The similarity of cellular processes in S. cerevisiae to those in higher eukaryotes and its accessibility to a wide range of experimental techniques have made this yeast a popular model for studying chronological aging of metazoan cells [1][2][3][4]. Chronological aging of S. cerevisiae is typically studied in aerobic batch cultures, in which growth arrest and quiescence are triggered by exhaustion of the available carbon sources in the growth medium [5][6]. Survival of individual yeast cells in such non-growing, stationary-phase (SP) cultures is then taken as a measure for their chronological lifespan (CLS). Over the past decade, studies on SP yeast cultures have contributed to our understanding of cellular mechanisms involved in aging, and several underlying cellular mechanisms were also found in higher eukaryotes [7].
–
Calorie restriction has been shown to extend lifespan in organisms ranging from yeast to man, with studies on many organisms pointing at an important role of nutrient-signaling cascades [8]. Turn-over of damaged macromolecules, and in particular proteins, has similarly been identified as a key process in aging in many organisms [9]. A third universal factor implicated in aging is respiration and, in particular, the associated formation of reactive oxygen species (ROS), which has been shown to enhance aging-related cellular deterioration in many organisms [10]. However, ROS have also been implicated in beneficial effects. In particular, mild ROS stress has been proposed to contribute to CLS extension by inducing stress-resistance genes, a phenomenon known as hormesis [11][12]. Similarly, increased mitochondrial respiration and ROS production rates in calorie-restricted yeast cultures have been linked to CLS extension [13][14][15].
–
ROS generation is not necessarily the only mechanism by which respiration and oxygen can affect CLS. In aerobic, glucose-grown batch cultures of S. cerevisiae, a fast and predominantly fermentative growth phase on glucose is followed by a second, respiratory growth phase in which the fermentation products ethanol and acetate are consumed [16]. This second growth phase, known as post-diauxic phase, is characterized by slow growth. During the post-diauxic phase, genes involved in SP are already expressed at an elevated level, as well as some features associated with SP cultures, such as increased stress resistance [6]. In anaerobic cultures of S. cerevisiae, the absence of oxygen prevents a respiratory post-diauxic growth phase. Instead, a phase of fast, fermentative exponential growth on glucose is immediately followed by SP, in which maintenance of viability and cellular integrity depends on metabolism of storage compounds. S. cerevisiae cells can contain two types of storage polymers: the storage carbohydrates trehalose and glycogen, and fatty acids, which are mostly stored in the form of di- and triacylglycerol esters [17][18][19]. In the absence of oxygen, yeast cells cannot catabolize fatty acids by β-oxidation and, moreover, conversion of storage carbohydrates via alcoholic fermentation yields 5-8 fold less ATP than their respiratory dissimilation [20].
–
Previous studies on the role of respiration in aging were predominantly based on the use of respiration-deficient S. cerevisiae mutants (e.g. ρ0 strains and other mutants) [21][22][23][24][25] and respiratory inhibitors [14]. These approaches, however, have several drawbacks. Firstly, mitochondria are not only involved in respiration, but also in essential anabolic reactions (e.g., assembly of iron-sulfur complexes, amino acid biosynthesis and long-chain lipid biosynthesis [26]). Studies on petite or ρ0 mutants may therefore cause unwanted ‘side-effects’ resulting from the absence or inefficiency of mitochondrial processes, rather than from direct effects of oxygen or respiration on aging. For example, the absence of mitochondrial DNA influences crosstalk between these organelles and the nucleus [27]. Furthermore, inhibition of respiration may result in reduced ROS levels [14], but can also result in ROS accumulation [28], depending on the intervention chosen. In addition, ROS may still be produced by other oxygen-consuming processes in yeast, such as disulfide-bond formation during oxidative protein folding [29].
–
Surprisingly, while S. cerevisiae is unique among yeasts and eukaryotes for its ability to grow fast under fully aerobic as well as strictly anaerobic conditions [30], this ability has not been used to systematically investigate the impact of oxygen availability on entry into SP, on longevity and on robustness. The goal of the present study was therefore to investigate the impact of oxygen availability on yeast physiology in SP cultures. More specifically, we investigated whether the post-diauxic phase and respiratory mobilization of storage compounds in aerobic cultures affects CLS and thermotolerance during SP. To this end, aerobic and anaerobic bioreactor batch cultures of S. cerevisiae were grown into SP and subjected to detailed physiological and transcriptome analyses.
RESULTS
Anaerobicity reduces chronological lifespan and stress resistance in stationary phase cultures
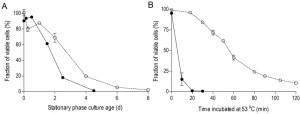
To investigate the impact of oxygen availability on chronological lifespan in SP cultures of S. cerevisiae, survival kinetics were analyzed during SP in aerobic and anaerobic, glucose-grown bioreactor cultures. In aerobic cultures, the percentage of cells capable of colony formation on complex-medium agar plates typically decreased to ca. 2% in the 8 days following onset of SP, i.e. after exhaustion of all exogenous carbon sources including ethanol and organic acids (Figure 1A). Viability of anaerobic cultures decreased much faster, reaching values below 1% within 4.5 days after the onset of SP, that is after all exogenous glucose was consumed (Figure 1A).
Increased thermotolerance is a well-documented characteristic of SP cultures of S. cerevisiae [31][32]. Indeed, half of the cells in samples from aerobic, early-SP cultures survived a 60-min exposure to 53°C. Notably, up to 20 minutes incubation at 53°C hardly affected viability, suggesting that cells were well capable of repairing heat-induced damage during this period. In contrast, fewer than 20% of the cells from anaerobic early-SP cultures survived a 10-min incubation at this temperature (Figure 1B). Implementation of anaerobic conditions during sampling and heat-shock assays did not significantly affect this difference, indicating that heat-induced loss of viability was not influenced by exposure of anaerobically grown cells to oxygen during the assays (data not shown). Furthermore, washing of cells prior to the heat-shock experiments did not influence heat-shock resistance, indicating that the presence of low (< 1 g/L) ethanol concentrations in the assays did not cause the low thermotolerance of cells from anaerobic SP cultures.
–
Oxygen availability strongly affects the transcriptome of SP cultures
In aerobic yeast cultures, entry into SP is accompanied by a range of physiological changes that enhance survival in harsh, nutrient-poor environments [31]. This adaptation coincides with a vast transcriptional reprogramming [33][34][35] that includes up-regulation of genes involved in resistance mechanisms to a wide array of stresses. Currently, no transcriptome data are available in the literature on anaerobic SP cultures of S. cerevisiae.
–
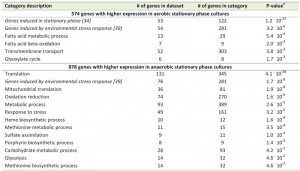
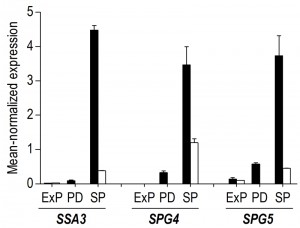
A transcriptome analysis, performed on culture samples taken 4 h after the onset of SP, revealed that a quarter of the yeast genome (1452 genes, Supplemental Table S1) was differentially expressed (fold-change cut-off of 2.0 and adjusted P-value below 0.05) in aerobic and anaerobic SP cultures. Among these genes were several genes known to be regulated by the heme and oxygen dependent transcription factors Hap1 and Rox1 [36][37][38]. Approximately 40% of the differentially expressed genes (574 genes, Supplemental Table S1) were transcribed at higher levels in the aerobic SP cultures. This gene set showed a strong overrepresentation of genes involved in fatty acid metabolism and, in particular, in β-oxidation (Table 1). This set of genes was also strongly enriched for genes that were up-regulated during SP in previous studies performed in shake flasks [34] (Table 1). Examples included the SP-genes SPG1, SPG3, SPG4, SPG5, and SSA3, which encodes a stress-induced ATPase. Furthermore, a significant number of genes (54) induced by the environmental stress response [39] was expressed at higher levels in aerobic SP cultures than in their anaerobic counterparts (Table 1).
Most of the genes that were differentially expressed in aerobic and anaerobic SP cultures (60%, 878 genes, Supplemental Table S1) showed a higher transcript level under anaerobic conditions. This gene set showed an overrepresentation of genes that were previously shown to be expressed at high levels in anaerobic yeast cultures and which were therefore not necessarily related to SP. This subgroup included genes involved in heme synthesis [38] and members of the multi-gene seripauperin family [40] (Table 1 and Supplemental Table S2). Interestingly, a strong overrepresentation (131 out of 345 genes) was found for the GO category ‘translation’ (Table 1). This subset included many genes encoding cytosolic and mitochondrial ribosomal proteins (65 and 24 genes respectively). Furthermore, several genes involved in carbohydrate metabolism, including glycogen metabolism, were expressed at higher levels in anaerobic SP cultures (Table 1). Finally, the set of 878 genes with higher expression in anaerobic SP cultures showed a very strong overrepresentation of genes induced by the environmental stress response [39].
–
Anaerobicity negatively affects the energy status of SP cultures
Use of oxygen as an electron acceptor for respiration enables oxidative phosphorylation. As a consequence, ATP yields on glucose, glycogen and trehalose in respiratory cultures can be up to ca. 8-fold higher than in anaerobic, fermentative cultures [20][41]. Furthermore, since anaerobic yeast cultures cannot catabolize fatty acids, their use as energy reserves is restricted to aerobic cultures. To investigate the impact of storage metabolism and energy status on the short CLS of anaerobic SP cultures, cellular contents of storage materials and adenylate energy charge (a measure for the energetic status of living cells [42]) were analysed in aerobic and anaerobic SP cultures.
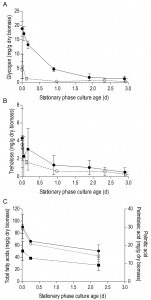
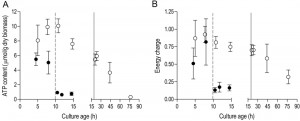
In aerobic cultures, intracellular pools of trehalose and glycogen were depleted within 1 day after entry into SP (Figure 2A and 2B). Cellular contents of the fatty acids palmitic and palmitoleic acid, also decreased during aerobic SP, but at a much slower rate than the storage carbohydrates and approached 6% of the total fatty acids content. This level is close to the membrane-associated fatty acid content previously reported in S. cerevisiae [43], indicating that yeast cells had exhausted most of their reserve lipids after 2 days in SP (Figure 2C). Together with the increased expression of genes involved in β-oxidation in aerobic SP cultures (Table 1), this observation indicates that aerobic SP cells use part of their fatty acids as an endogenous carbon and energy source. In aerobic cultures, the adenylate energy charge was 0.70 (± 0.08) and the intracellular ATP concentration was 5.45 (± 0.76) mM at the onset of SP. These results are in good agreement with published data [44][45]. In the days after the onset of SP, both parameters gradually decreased. Two full days after the onset of SP, the adenylate energy charge was still above 0.25 (Figure 3).
Trehalose and glycogen are the only known carbon and energy reserves in anaerobic S. cerevisiae cultures. The initial trehalose content and utilization profile in anaerobic SP cultures strongly resembled those observed in aerobic cultures. The initial glycogen concentration in anaerobic SP cultures was ca. four-fold higher than in aerobic SP cultures. Nevertheless, intracellular glycogen was exhausted after 2 days in SP (Figure 2A and 2B). At the onset of SP, intracellular ATP concentration and adenylate energy charge of anaerobic SP cultures were already lower than in aerobic cultures. Moreover, they decreased to very low levels within half a day after the onset of SP (Figure 3).
–
Different entry trajectories into SP in aerobic and anaerobic cultures
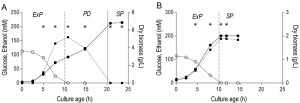
The results described above reveal clear differences in transcriptome, energy status, thermotolerance and CLS of aerobic and anaerobic SP cultures. Some of these parameters already differed at the onset of SP, indicating the importance of different ‘entry trajectories’ of aerobic and anaerobic cultures into SP. A major difference between aerobic and anaerobic batch cultures is the absence, in the latter, of a respiratory post-diauxic phase, in which ethanol and minor fermentation products acetate and glycerol are consumed. Growth in the post-diauxic phase, in which metabolism is completely respiratory, is slower than during the preceding glucose phase [46]. In this study, the maximum specific growth rate of anaerobic cultures (0.31 ± 0.01 h-1) during the glucose phase was lower than that of aerobic cultures (0.39 ± 0.02 h-1, Figure 4). In aerobic cultures, the specific growth rate during the post-diauxic phase (0.10 ± 0.01 h-1) was ca. four-fold lower than during the fast growth phase on glucose. As a consequence, the specific growth rate in the hours preceding the onset of SP was ca. three-fold lower in aerobic cultures than in anaerobic cultures. To investigate whether the post-diauxic phase may have ‘conditioned’ aerobic cultures for entry into SP, analysis of aerobic and anaerobic batch cultures was extended to include the growth phases that precede SP.
A much higher thermotolerance in aerobic cultures was already evident in the exponential growth phase and further increased during the post-diauxic phase, to reach a maximum upon entry into SP (Figure 5). Conversely, thermotolerance of anaerobic cultures did not increase during entry into SP and, consequently, remained much lower than that of aerobic cultures (Figure 5).
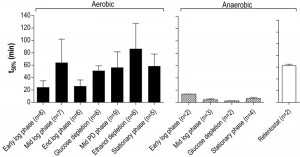 | FIGURE 5: Thermotolerance of aerobic and anaerobic cultures during different growth phases. Thermotolerance of cells during different growth phases of aerobic (black bars) and anaerobic (hatched bars) batch cultures of S. cerevisiae (Figure 4). The white bar depicts the thermotolerance of S. cerevisiae grown for 8 days in anaerobic retentostats [20]. Thermotolerance was assayed by monitoring viability during incubation at 53°C and is shown as the incubation time resulting in a 50% decrease in viability (t50) (see Materials and Methods for more details). The number of independent culture replicates for each of the growth phases is denoted on the x-axis labels. |
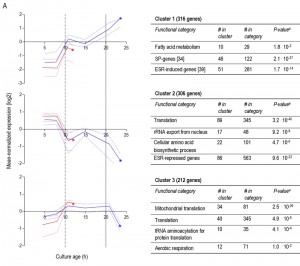
To further compare the different ‘entry trajectories’ into SP of aerobic and anaerobic batch cultures, transcriptome analyses were performed at different time points during exponential phase, post-diauxic phase (aerobic cultures only) and SP. Genes were grouped in 9 clusters, based on their time-dependent expression profiles in aerobic and anaerobic cultures (Figure 6). A full dataset is available in Supplemental Data (Tables S3 and S4).
–
Less than one tenth (126 of 1452) of the genes that were differentially expressed in aerobic and anaerobic SP cultures (Table 1) already showed corresponding differences during the mid-exponential growth phase in aerobic and anaerobic cultures. For over half (834 of 1452) of the differentially expressed genes in SP cultures, the differences rose after glucose exhaustion, i.e. during the post-diauxic phase in aerobic cultures (Figure 6A, clusters 1-3). Cluster 1 comprises genes whose expression increased during the aerobic and anaerobic exponential growth phases, with a further increase during the aerobic post-diauxic phase (Figure 6A). Genes involved in fatty acid catabolism were overrepresented in this cluster, as well as genes that were previously shown to be induced in aerobic SP (including SPG1, SPG3, SPG4, SPG5 and Hsp70–family–member SSA3 [34]) (Figures 6A and 7). Genes in cluster 2 showed similar transcript levels during the aerobic and anaerobic exponential growth phases on glucose. However, due to a pronounced decrease in expression during the post-diauxic phase in aerobic cultures, expression levels were higher in anaerobic SP cultures than in aerobic SP cultures. This cluster was markedly enriched for genes involved in amino acid synthesis and translation (Figure 6A), suggesting that a down-regulation of protein synthesis occurred during the post-diauxic phase. Cluster 3 comprised of genes whose transcript levels increased during the exponential phase of both aerobic and anaerobic cultures but, subsequently, only decreased in aerobic SP cultures. Cluster 3 showed an overrepresentation of genes involved in mitochondrial translation and respiration (Figure 6A).
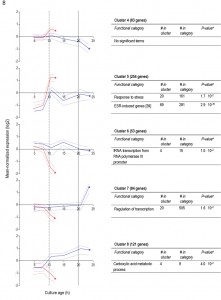
For 595 of the ‘oxygen-responsive in SP’ genes listed in Table 1, differences in expression occurred already upon glucose exhaustion (Figure 6B, cluster 4-8). Genes in Cluster 5, characterized by a specific up-regulation upon entry into anaerobic SP, showed an overrepresentation of stress-responsive genes (Figure 6B, cluster 5). Several of these are known to be specifically expressed under anaerobic conditions (e.g., the cell-wall mannoprotein-encoding gene DAN4 and members of the seripauperin family [40]), but cluster 5 also included heat-shock genes whose expression is not specifically linked to anoxic conditions (e.g. HSP30 and SSA4). Genes that showed a specific downregulation during anaerobic SP, but a constant (Figure 6B, cluster 6) or increased expression in aerobic SP (Figure 6B, clusters 7 and 8) showed an overrepresentation of genes involved in transcription-related processes and carboxylic acid metabolism. The latter of which plays a role in the respiration of exogenous carboxylic acids during the post-diauxic phase (Figure 6B, cluster 8). All 23 genes whose transcript levels were higher under anaerobic conditions, irrespective of the growth phase (Supplemental Table S4), were previously described to be upregulated under anaerobic conditions. The majority (14) of these genes belonged to the seripauperin family [40].
Two clusters (Figure 6, cluster 1 and 5) comprised genes whose transcript levels increased in aerobic as well as in anaerobic batch cultivation, but to different final levels. These clusters were enriched for genes induced by the environmental stress response [39]. The extreme differences in thermotolerance of aerobic and anaerobic SP cultures (Figure 5) were therefore only partially mirrored at the transcript level, indicating that factors other than transcriptional reprogramming contribute to these differences.
DISCUSSION
This study demonstrates a strong impact of oxygen availability on chronological lifespan and stress tolerance in SP batch cultures of S. cerevisiae and, thereby, confirms and extends earlier observations on its physiology in aerobic and anaerobic cultures [13][19][47]. The CLS of anaerobically grown SP cultures was much shorter than that of their aerobic counterparts and an even more dramatic difference was observed for thermotolerance. As will be discussed below, these differences involved a different conditioning of aerobic and aerobic cultures during the growth phases preceding SP, as well as energetic constraints imposed on yeast cells in anaerobic SP cultures.
–
The post-diauxic growth phase enables transcriptional conditioning of aerobic yeast cultures for stationary phase
Our transcriptome data revealed that 57% of the transcriptional differences between aerobic and anaerobic SP cultures originated from transcriptional reprogramming during the aerobic post-diauxic growth phase. Although several well-known hallmark transcripts of SP cultures, previously identified in (semi-) aerobic shake-flask cultures, such as SPG4, SPG5 and SSA3 [34], showed increased levels in anaerobic SP cultures, their levels did not reach those observed in aerobic cultures (Figure 7). Moreover, many genes involved in biosynthesis were strongly down-regulated during the aerobic post-diauxic phase and SP, but retained expression levels close to those in the exponential growth phase in anaerobic cultures (Figure 6). These transcriptome data are consistent with the hypothesis that the post-diauxic phase in aerobic cultures conditions cells for entry into SP and that, conversely, absence of a post-diauxic phase prevents anaerobic batch cultures from adequately adapting to SP and starvation.
–
Hormesis could potentially explain the difference in robustness between aerobic and anaerobic cultures. Indeed respiration can generate low levels of ROS and thereby induce stress tolerance via increased expression of stress tolerance genes [13][21]. However, among a set of 22 genes encoding enzymes involved in ROS-protective mechanisms [48], including the superoxide dismutase genes SOD1 and SOD2, whose expression is strongly upregulated during exposure to ROS [13][39], only the peroxisomal catalase CTA1 was higher expressed in aerobic SP cultures. These findings argue against a dominant role of ROS-based hormesis in the acquisition of increased robustness by aerobic SP cultures.
–
Caloric restriction: a key factor in conditioning yeast cells for stationary phase and starvation
Thermotolerance is negatively correlated with specific growth rate in S. cerevisiae as Lu et al. (2009) demonstrated in nutrient-limited chemostat cultures [49]. Although these authors did not evaluate the impact of specific growth rate on CLS nor investigate anaerobic growth, they showed that the negative correlation between thermotolerance and growth rate also held in a respiratory-deficient S. cerevisiae strain [49]. Our data are fully consistent with the hypothesis that the strong reduction of specific growth rate (from 0.39 to 0.10 h-1) during transition from fast exponential growth on glucose to the post-diauxic phase in aerobic batch cultures could similarly trigger increased thermotolerance and extended CLS during the starvation phase. We have also recently shown that a gradual decrease of the specific growth rate to near-zero values in glucose-limited retentostats [50] yielded yeast cells with a thermotolerance that is as high as that of aerobic SP cultures (Figure 5), and with an even longer CLS during subsequent starvation [20]. The transcriptional reprogramming observed in these anaerobic severely calorie-restricted cultures [51] strongly resembled the transcriptome changes observed in the present study for aerobic cultures entering SP and proteome analysis showed increased levels of proteins involved in stress resistance [52]. Deletion of Rim15, a kinase under control of several nutrient signaling pathways [53], strongly reduced the acquisition of robustness in both anaerobic and aerobic calorie-restricted cultures [54][55], suggesting a strong role for nutrient signaling independent of oxygen availability.
–
The present study, combined with our previous retentostat studies, therefore clearly demonstrates that prior conditioning by a period of caloric restriction (e.g. by slow growth during the aerobic post-diauxic phase or in extremely glucose-limited cultures) is a prerequisite for acquisition of a prolonged CLS by non-growing, starving cultures of S. cerevisiae. This conclusion, which supports earlier proposals based on starvation experiments by Thomsson et al. [47][56], has important implications for the design and interpretation of yeast studies on chronological aging, for example when such studies involve mutants that are impeded in energy metabolism.
–
The low thermotolerance of exponentially growing anaerobic cultures does not correlate with expression of heat shock genes
Although both aerobic and anaerobic, non-growing yeast cultures can acquire a similar thermotolerance by an appropriate preceding conditioning phase, a drastic difference was observed in the thermotolerance of exponentially growing aerobic and anaerobic cultures (Figure 5). The similar adenylate energy charge and intracellular ATP concentrations of aerobic and anaerobic cultures during exponential growth on glucose appear to rule out cellular energy status as a major cause of this difference. Since the high temperature (53°C) during the thermotolerance assays precludes de novo synthesis of mRNA or protein synthesis [57], this difference must already be expressed in the batch cultures themselves. Transcript levels of genes that were previously implicated in heat-shock resistance (including HSP genes [58] such as HSP104 [59], HSP26 [60], HSP12 [61], SSA3 [62]) were similar during the exponential growth phase on glucose in aerobic and anaerobic cultures. Moreover, of 59 genes identified as essential for heat-shock survival by Gibney and coworkers [57], only one gene was differentially expressed, LIA1, and showed a higher transcript level in anaerobic cultures.
–
Oxygen availability strongly influences sterol and unsaturated fatty acid composition of yeast cells [19], especially because these compounds have to be added to growth media as anaerobic growth factors [43]. These differences in membrane composition might partially explain the observed differences in thermotolerance between aerobically and anaerobically grown S. cerevisiae cells. The hypothesis that membrane composition is a key determinant in thermotolerance of S. cerevisiae [63][64] is consistent with a recent study, in which the acquisition of increased thermotolerance by laboratory-evolved strains was shown to be caused by changes in their sterol composition [65].
–
A low energy status of anaerobic SP cultures limits metabolic flexibility
Consistent with earlier reports [19][66], anaerobic batch cultures of S. cerevisiae displayed a substantially higher glycogen content than aerobic cultures. However, after the onset of SP, anaerobic cultures showed a much faster decrease of the adenylate energy charge. This difference can be attributed to several factors. Firstly, since S. cerevisiae cannot derive metabolic energy from lipids and amino acids in the absence of oxygen [67], anaerobic cultures are entirely dependent on glycogen and trehalose as energy storage compounds and anaerobic catabolism of these storage carbohydrates yields less ATP than respiration. The estimated ATP synthesis rate from anaerobic glycogen dissimilation of ca. 5 µmol per g biomass dry weight per hour during the first day in SP (based on a maximum ATP yield of 3 ATP per glucose residue [20]), was two orders of magnitude lower than the cellular ATP demand for maintenance estimated from chemostat and retentostat cultures (mATP = 1 mmol ATP per g biomass dry weight per hour [50]). A similar extreme reduction of ATP turnover rates was observed when anaerobic retentostat cultures were switched to carbon starvation [20]. Together, these observations indicate that an extremely low ATP turnover is an intrinsic feature of anaerobic, starving yeast cultures. In addition to this extreme low ATP-turnover, it is even conceivable that the apparent inability of anaerobic batch cultures to efficiently down-regulate energy-consuming processes, including protein synthesis, the single most expensive biosynthetic process in living cells [68][69], may have exacerbated the fast decline of their energy status after entry into SP (Figure 3).
–
The low energy status of cells may at the same time have put strong constraints on these energy consuming processes. Proteome analyses should reveal whether the increased transcription of HSP genes, implicated in thermotolerance, which took place late in the exponential growth phase (Figure 6), was too late to enable synthesis of the corresponding proteins before the decline in cellular energy status in anaerobic SP cultures. Such a scenario would explain the discrepancy between the oxygen-independent upregulation of these genes (with notable exception of SSA3) and the absence of increased thermotolerance in anaerobic SP cultures.
–
Taken together, the results from the present study indicate that, in the short time lapse between the moment at which anaerobic cultures sense that glucose reaches critically low levels and the actual exhaustion of glucose, they lack the time and resources to perform the energy-intensive remodeling of their transcriptome and proteome required to robustly face starvation. Our data are therefore entirely consistent with the notion that the low CLS and thermotolerance of anaerobic SP cultures, in comparison with aerobic cultures, is due to the absence of a proper conditioning phase and a limited metabolic flexibility due to a lower cellular energy status.
–
Outlook
Many studies in which SP yeast cultures are used as a model system to investigate aspects of aging, still rely on shake-flask cultures. Due to their low and poorly controlled oxygen-transfer capacity, the aeration status of shake-flask cultures is generally unclear. The strong impact of oxygen availability on aging-related characteristics [14] underlines the value of controlled cultivation techniques, e.g. in bioreactors, including batch, chemostat and retentostat cultures [50][70][71][72] or flow-through cells [73], in yeast-based aging studies. In particular, the use of anaerobic cultures as a model offers interesting possibilities to clarify the role of respiration and ROS in aging, apoptosis and longevity.
–
The short life span and low robustness of anaerobic SP cultures of S. cerevisiae is directly relevant for industrial applications. Robustness of SP cultures is especially important for processes in which biomass from anaerobic batch cultures is recycled, e.g. in industrial bioethanol production and beer brewing [74][75]. Clearly, results from (semi-)aerobic shake-flask cultures cannot be used to predict the performance of such anaerobic processes and improvement of robustness in these industrial processes will have to be based on studies in anaerobic systems.
–
Saccharomyces yeasts have the capability, rare among eukaryotes, to grow fast in the complete absence of oxygen and it is often assumed that they are well adapted to anaerobic environments [30][76][77]. While the natural habitat of S. cerevisiae is still a matter of debate [77], lower biomass concentrations frequently encountered in natural environments combined with the low affinity of yeast glucose transporters [78] may lead to a transition into SP that is sufficiently slow to enable acquisition of longevity and robustness under anaerobic conditions. Further research is therefore needed to investigate the ecological relevance of this laboratory study.
MATERIALS AND METHODS
Strains and cultivation
The prototrophic Saccharomyces cerevisiae strain CEN.PK113-7D (MATa MAL2-8c SUC2) used in this study is a congenic member of the CEN.PK family [79][80]. Stock cultures were grown at 30°C in shake flasks containing yeast extract (1% w/v), peptone (2% w/v) and dextrose (2% w/v) (YPD) medium. Glycerol, final concentration 20% (v/v), was added to overnight cultures and 1 mL aliquots were stored at -80°C.
–
Previously described synthetic medium [81] was used with 20 g/L glucose as sole carbon-source and 0.2 g/L antifoam Emulsion C (Sigma, St. Louise, USA). In case of anaerobic cultivations, the medium was supplemented with anaerobic growth factors ergosterol (10 mg/L) and Tween 80 (420 mg/L) dissolved in ethanol. Inocula for batch fermentations consisted of 100 mL yeast culture grown overnight to an OD660 of 4 in synthetic medium with 20 g/L glucose. Aerobic and anaerobic batch fermentations were carried out at 30°C in 2 L bioreactors (Applikon, Schiedam, The Netherlands), with a working volume of 1.4 L. Cultures were stirred at 800 rpm and sparged at a flow-rate of 700 mL/min with either dried air or nitrogen gas (< 10 ppm oxygen, Linde Gas Benelux, The Netherlands). The bioreactors were equipped with Norprene tubing (Saint-Gobain Performance Plastics, Courbevoie, France) and Viton O-rings (Eriks, Alkmaar, The Netherlands) to minimize diffusion of oxygen. During aerobic cultivations, dissolved oxygen levels remained above 40% of the initial saturation level as measured by Clark electrodes (Mettler Toledo, Greifensee, Switzerland). The culture pH was maintained at 5.0 by automatic addition of 2 M KOH and 2 M H2SO4.
–
Analysis of biomass, metabolites, substrate and exhaust gas
Biomass concentration as culture dry weight was determined as described previously [82].
–
For substrate and extracellular metabolite concentration determination, culture supernatants were obtained by centrifugation of culture samples (3 min at 20.000 g) and analysed by high-performance liquid chromatograph (HPLC) analysis on a Waters Alliance 2690 HPLC (Waters, Milford, MA) equipped with a Bio-Rad HPX 87H ion exchange column (BioRad, Veenendaal, The Netherlands), operated at 60°C with 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml/min. Detection was by means of a dual-wavelength absorbance detector (Waters 2487) and a refractive index detector (Waters 2410).
–
The exhaust gas from batch cultures was cooled with a condenser (2°C) and dried with a PermaPure Dryer (model MD 110-8P-4; Inacom Instruments, Veenendaal, the Netherlands) prior to online analysis of carbon dioxide and oxygen with a Rosemount NGA 2000 Analyser (Baar, Switzerland).
–
Colony forming units
To determine culture viability, small aliquots of culture broth were taken from the reactor and cells were counted on a Z2 Coulter Counter (Beckman Coulter, Woerden, Netherlands) equipped with a 50 µm orifice (Multisizer II, Beckman Coulter, Woerden, Netherlands). Cells were diluted in 0.1% peptone and 100 µL suspensions containing approximately 30, 300 and 3000 cells were plated on yeast extract (1% w/v), peptone (2% w/v) and dextrose (0.5% w/v)) (YPD) agar plates and incubated at 30°C for at least 3 days before counting the colonies. CFU was calculated as the number of colonies formed divided by the number of plated cells.
–
Thermotolerance assay
Cells from culture broth were counted with a Z2 Coulter Counter and diluted in pre-warmed (53°C) isotone diluent II (Beckman Coulter, Woerden, Netherlands) to yield 50 mL cell suspensions with a density of 1∙107 cells/mL. Cell suspensions were incubated in a waterbath at 53°C and 4 mL aliquots were sampled in 10 min intervals. Samples were cooled on ice and assayed for viability using the FungaLight 5-CFDA, AM (acetoxymethyl ester 5-carboxyfluorescein diacetate)/propidum iodide (PI) yeast viability kit (Invitrogen, Carlsbrad, CA) by counting 10,000 cells on a Cell Lab Quanta SC MPL flow cytometer (Beckman Coulter, Woerden, Netherlands) as described previously [20]. 5-CFDA, AM is a cell-permeant substrate for intracellular non-specific esterase activity. Hydrolytic cleavage of the lipophilic blocking acetoxymethyl and diacetate groups of 5-CFDA, AM results in a green fluorescent signal in metabolically active cells. Propidium iodide intercalates with DNA in cells with a compromised cell membrane, which results in red fluorescence. Cells stained with PI were considered dead cells. For each independent sample, the t50 value (the time after which 50% of the initial viable population was dead) was estimated by fitting the viability data with a sigmoidal dose-response curve in Graphpad Prism 4.03. Both measurements of viability, i.e. metabolic activity based on 5-CFDA, AM and membrane integrity based on PI gave similar t50-values, therefore only results based on PI are shown.
–
Storage carbohydrate measurements
1.2 mL culture broth was quenched in 5 mL of cold methanol (-40°C) using a rapid sampling setup described previously [83], mixed and subsequently pelleted (4,400 g, 5 min) at -19°C. Cells were washed with 5 mL of cold methanol (-40oC) and pellets stored at -80°C. Pellets were resuspended in Na2CO3 (0.25 M) and further processed according to a previously described procedure [84].
–
Fatty acids measurements
Culture volumes corresponding to 50 mg biomass were sampled on ice, centrifuged (10,000 g, 10 min at 4°C), washed twice, resuspended in 5 ml ice-cold water and stored at -20°C. Lipid extraction was performed as described previously [85]. Aliquots of 0.15 mL were added to 15 mL tubes and 1.5 mL of a mixture of concentrated HCl and 1-propanol (1:4) and 1.5 mL of dichlorethane were added. 400 μg of myristic acid (a 15:0 fatty acid) was included as internal standard. Samples were incubated at 100°C for 2 h. Subsequently, 3 mL of water was added to cooled samples. 1 mL of the organic phase was filtered over water-free sodium sulfate into GC vials. The fatty acid propyl esters in the organic phase were analyzed by gas chromatography (model 6890N, Agilent, U.S.A.) using a DB-wax column (length, 30 m; inside diameter, 0.25 mm; film thickness, 0.25 μm; J&W Scientific, Folsom, CA) and helium as the carrier gas. The sample volume was 1 μL, and the split was set to 1:20. The injection temperature was 230°C, and the following temperature gradient was used: 120°C at the start, increasing at a rate of 10°C/min up to 240°C, and then 240°C for 8 min. The fatty acid propyl esters were detected using a flame ionization detector at 250°C.
–
Analysis of intracellular adenosine-phosphate concentrations
Samples for internal metabolite analysis were obtained by rapid sampling [83]. 1.2 mL of culture broth was rapidly quenched into 5 mL of 100% methanol, pre-cooled to -40°C. Samples were washed with cold methanol and extracted with boiling ethanol. Intracellular AMP and ADP were determined enzymatically, using a previously described assay based on myokinase, pyruvate kinase and lactate dehydrogenase reactions [86]. Assays were performed in white, flat bottom 96-well microtiter plates (Corning Inc., USA). NADH fluorescence was measured in a TECAN GENios Pro microtiterplate reader (Tecan, Männedorf, Switzerland) as previously described [83]. Intracellular ATP was also assessed enzymatically. The assay contained 115 mM triethanolamine (pH 7.6), 11.5 mM MgSO4x7H2O, 1.15 mM NADP+ per well including sample extract, total volume was 205 µL per well. The reaction to measure ATP was initiated by adding 12 mM glucose and 30 U hexokinase (Sigma-Aldrich Chemie B.V, Zwijndrecht, The Netherlands). Assays were performed in black, flat bottom 96-well microtiter plates (Corning Inc., USA). NADPH fluorescence was measured in a TECAN GENios Pro microtiterplate reader. The adenylate energy charge was calculated according to the previously described [42] equation:
–
 |
–
Transcriptome analysis
Independent duplicate aerobic and anaerobic batch cultures were sampled at six and four different time points respectively (see Figure 4A and 4B) for microarray analysis, resulting in a total dataset of 20 microarrays. Sampling from batch cultures for transcriptome analysis was performed using liquid nitrogen for rapid quenching of mRNA turnover [87]. Prior to RNA extraction, samples were stored in a mixture of phenol/chloroform and TEA buffer at -80°C. Total RNA extraction, isolation of mRNA, cDNA synthesis, cRNA synthesis, labelling and array hybridization was performed as previously described [88]. The quality of total RNA, cDNA, aRNA and fragmented aRNA was checked using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Hybridization of labelled fragmented aRNA to the microarrays and staining, washing and scanning of the microarrays was performed according to Affymetrix instructions (EukGE_WS2v5).
–
The 6383 yeast open reading frames were extracted from the 9335 transcript features on the YG-S98 microarrays. To allow comparison, all expression data were normalized to a target value of 240 using the average signal from all gene features [89]. The microarray data used in this study are available via GEO series accession number GSE69485. To eliminate variation in genes that are not expressed, genes with expression values below 12 were set to 12 and the gene features for which the maximum expression was below 20 for all 20 arrays were discarded [51]. The average deviation of the mean transcript data of replicate batches was ca. 11%, similar to the reproducibility usually observed in replicate steady state chemostat cultures [90]. The expression of housekeeping genes ACT1, PDA1, TFC1, ALG9, TAF10 and UBC6 [91] remained stable for both conditions and all sample points (average coefficient of variation 17% ± 5%).
–
To identify genes that were differentially expressed between aerobic and anaerobic SP cultures, a pairwise comparison was performed between aerobic samples taken at time point 6 (Figure 4A) and anaerobic samples taken at time point 4 (Figure 4B) as previously described [89]. Similarly, genes differently expressed during growth on glucose under aerobic or anaerobic conditions were identified through a pairwise comparison of aerobic and anaerobic samples taken at time point 1 (Figure 4A and 4B). Differences with adjusted P-values lower than 0.05 and a fold difference of 2 or higher were considered statistically significant. Time-dependent expression profiles of selected genes were clustered according to optimal k-means clustering using positive correlation as distance metric (Expressionist Pro version 3.1, Genedata, Basel, Switzerland) resulting in an optimal number of 9 clusters. For display of time-dependent transcript levels, expression values were normalized per gene by dividing single expression values by the average expression value of both conditions and all time points. Mean values of these average-normalized values for all genes in each cluster are shown, as well as the first and third quartile of average-normalized values.
–
Gene expression clusters were analysed for overrepresentation of functional annotation categories of the Gene Ontology (GO) database (http://www.geneontology.org/), based on a hypergeometric distribution analysis tool [92]. Additional categories describing genes expressed in SP cultures [34], genes commonly induced by several environmental stresses [39] or essential for heat-shock survival [57] were extracted from the respective references.
References
- A. Denoth Lippuner, T. Julou, and Y. Barral, "Budding yeast as a model organism to study the effects of age", FEMS Microbiology Reviews, vol. 38, pp. 300-325, 2014. http://dx.doi.org/10.1111/1574-6976.12060
- V. Longo, G. Shadel, M. Kaeberlein, and B. Kennedy, "Replicative and Chronological Aging in Saccharomyces cerevisiae", Cell Metabolism, vol. 16, pp. 18-31, 2012. http://dx.doi.org/10.1016/j.cmet.2012.06.002
- M. Breitenbach, S. Jazwinski, P. Laun, and . editors, "Aging research in yeast", Springer Netherlands, Dordrecht., 2012.
- M.G. Mirisola, R.J. Braun, and D. Petranovic, "Approaches to study yeast cell aging and death", FEMS Yeast Research, vol. 14, pp. 109-118, 2013. http://dx.doi.org/10.1111/1567-1364.12112
- V. Longo, and P. Fabrizio, "Chronological aging in Saccharomyces cerevisiae", In: Breitenbach M, Jazwinski SM, Laun P, editors. Aging Research in Yeast. Springer Netherlands, Dordrecht; pp 101-121., 2012.
- J.V. Gray, G.A. Petsko, G.C. Johnston, D. Ringe, R.A. Singer, and M. Werner-Washburne, "“Sleeping Beauty”: Quiescence inSaccharomyces cerevisiae", Microbiology and Molecular Biology Reviews, vol. 68, pp. 187-206, 2004. http://dx.doi.org/10.1128/mmbr.68.2.187-206.2004
- P. Fabrizio, and V.D. Longo, "The chronological life span of Saccharomyces cerevisiae", Aging Cell, vol. 2, pp. 73-81, 2003. http://dx.doi.org/10.1046/j.1474-9728.2003.00033.x
- D.A. Sinclair, "Toward a unified theory of caloric restriction and longevity regulation", Mechanisms of Ageing and Development, vol. 126, pp. 987-1002, 2005. http://dx.doi.org/10.1016/j.mad.2005.03.019
- A.G. Ryazanov, and B.S. Nefsky, "Protein turnover plays a key role in aging", Mechanisms of Ageing and Development, vol. 123, pp. 207-213, 2002. http://dx.doi.org/10.1016/s0047-6374(01)00337-2
- R.S. Balaban, S. Nemoto, and T. Finkel, "Mitochondria, Oxidants, and Aging", Cell, vol. 120, pp. 483-495, 2005. http://dx.doi.org/10.1016/j.cell.2005.02.001
- P. Ludovico, and W.C. Burhans, "Reactive oxygen species, ageing and the hormesis police", FEMS Yeast Research, vol. 14, pp. 33-39, 2013. http://dx.doi.org/10.1111/1567-1364.12070
- P.K. Sharma, V. Agrawal, and N. Roy, "Mitochondria-mediated hormetic response in life span extension of calorie-restricted Saccharomyces cerevisiae", AGE, vol. 33, pp. 143-154, 2010. http://dx.doi.org/10.1007/s11357-010-9169-1
- A. Mesquita, M. Weinberger, A. Silva, B. Sampaio-Marques, B. Almeida, C. Leão, V. Costa, F. Rodrigues, W.C. Burhans, and P. Ludovico, "Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H 2 O 2 and superoxide dismutase activity", Proceedings of the National Academy of Sciences, vol. 107, pp. 15123-15128, 2010. http://dx.doi.org/10.1073/pnas.1004432107
- A. Ocampo, J. Liu, E. Schroeder, G. Shadel, and A. Barrientos, "Mitochondrial Respiratory Thresholds Regulate Yeast Chronological Life Span and its Extension by Caloric Restriction", Cell Metabolism, vol. 16, pp. 55-67, 2012. http://dx.doi.org/10.1016/j.cmet.2012.05.013
- D.L. Smith, Jr, J.M. McClure, M. Matecic, and J.S. Smith, "Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins", Aging Cell, vol. 6, pp. 649-662, 2007. http://dx.doi.org/10.1111/j.1474-9726.2007.00326.x
- J.P. van Dijken, R.A. Weusthuis, and J.T. Pronk, "Kinetics of growth and sugar consumption in yeasts", Antonie van Leeuwenhoek, vol. 63, pp. 343-352, 1993. http://dx.doi.org/10.1007/BF00871229
- J. François, and J.L. Parrou, "Reserve carbohydrates metabolism in the yeastSaccharomyces cerevisiae", FEMS Microbiology Reviews, vol. 25, pp. 125-145, 2001. http://dx.doi.org/10.1111/j.1574-6976.2001.tb00574.x
- H. Kohei, and Y. Satoshi, "Regulatory role of phosphatidate phosphatase in triacylglycerol synthesis of Saccharomyces cerevisiae", Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism, vol. 796, pp. 110-117, 1984. http://dx.doi.org/10.1016/0005-2760(84)90244-3
- K. Wilson, and B.J. McLeod, "The influence of conditions of growth on the endogenous metabolism of Saccharomyces cerevisiae: effect on protein, carbohydrate, sterol and fatty acid content and on viability", Antonie van Leeuwenhoek, vol. 42, pp. 397-410, 1976. http://dx.doi.org/10.1007/BF00410171
- L.G. Boender, M.J. Almering, M. Dijk, A.J. van Maris, J.H. de Winde, J.T. Pronk, and P. Daran-Lapujade, "Extreme calorie restriction and energy source starvation in Saccharomyces cerevisiae represent distinct physiological states", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1813, pp. 2133-2144, 2011. http://dx.doi.org/10.1016/j.bbamcr.2011.07.008
- E.B. Tahara, F.M. Cunha, T.O. Basso, B.E. Della Bianca, A.K. Gombert, and A.J. Kowaltowski, "Calorie Restriction Hysteretically Primes Aging Saccharomyces cerevisiae toward More Effective Oxidative Metabolism", PLoS ONE, vol. 8, pp. e56388, 2013. http://dx.doi.org/10.1371/journal.pone.0056388
- S. Lin, M. Kaeberlein, A.A. Andalis, L.A. Sturtz, P. Defossez, V.C. Culotta, G.R. Fink, and L. Guarente, "Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration", Nature, vol. 418, pp. 344-348, 2002. http://dx.doi.org/10.1038/nature00829
- N.D. Bonawitz, M. Chatenay-Lapointe, Y. Pan, and G.S. Shadel, "Reduced TOR Signaling Extends Chronological Life Span via Increased Respiration and Upregulation of Mitochondrial Gene Expression", Cell Metabolism, vol. 5, pp. 265-277, 2007. http://dx.doi.org/10.1016/j.cmet.2007.02.009
- A.M. Aerts, P. Zabrocki, G. Govaert, J. Mathys, D. Carmona-Gutierrez, F. Madeo, J. Winderickx, B.P. Cammue, and K. Thevissen, "Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast", FEBS Letters, vol. 583, pp. 113-117, 2008. http://dx.doi.org/10.1016/j.febslet.2008.11.028
- M.H. Barros, F.M. da Cunha, G.A. Oliveira, E.B. Tahara, and A.J. Kowaltowski, "Yeast as a model to study mitochondrial mechanisms in ageing", Mechanisms of Ageing and Development, vol. 131, pp. 494-502, 2010. http://dx.doi.org/10.1016/j.mad.2010.04.008
- M. Breitenbach, M. Rinnerthaler, J. Hartl, A. Stincone, J. Vowinckel, H. Breitenbach-Koller, and M. Ralser, "Mitochondria in ageing: there is metabolism beyond the ROS", FEMS Yeast Research, vol. 14, pp. 198-212, 2014. http://dx.doi.org/10.1111/1567-1364.12134
- D.K. Woo, and R.O. Poyton, "The absence of a mitochondrial genome in rho0 yeast cells extends lifespan independently of retrograde regulation", Experimental Gerontology, vol. 44, pp. 390-397, 2009. http://dx.doi.org/10.1016/j.exger.2009.03.001
- J. Leadsham, G. Sanders, S. Giannaki, E. Bastow, R. Hutton, W. Naeimi, M. Breitenbach, and C. Gourlay, "Loss of Cytochrome c Oxidase Promotes RAS-Dependent ROS Production from the ER Resident NADPH Oxidase, Yno1p, in Yeast", Cell Metabolism, vol. 18, pp. 279-286, 2013. http://dx.doi.org/10.1016/j.cmet.2013.07.005
- E. Rosenfeld, and B. Beauvoit, "Role of the non‐respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae", Yeast, vol. 20, pp. 1115-1144, 2003. http://dx.doi.org/10.1002/yea.1026
- W. Visser, W.A. Scheffers, W.H. Batenburg-van der Vegte, and J.P. van Dijken, "Oxygen requirements of yeasts.", Applied and environmental microbiology, 1990. http://www.ncbi.nlm.nih.gov/pubmed/2082825
- M. Werner-Washburne, E. Braun, G.C. Johnston, and R.A. Singer, "Stationary phase in the yeast Saccharomyces cerevisiae.", Microbiological reviews, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8393130
- C. Allen, S. Büttner, A.D. Aragon, J.A. Thomas, O. Meirelles, J.E. Jaetao, D. Benn, S.W. Ruby, M. Veenhuis, F. Madeo, and M. Werner-Washburne, "Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures", The Journal of Cell Biology, vol. 174, pp. 89-100, 2006. http://dx.doi.org/10.1083/jcb.200604072
- K. Wanichthanarak, N. Wongtosrad, and D. Petranovic, "Genome-wide expression analyses of the stationary phase model of ageing in yeast", Mechanisms of Ageing and Development, vol. 149, pp. 65-74, 2015. http://dx.doi.org/10.1016/j.mad.2015.05.008
- M.J. Martinez, S. Roy, A.B. Archuletta, P.D. Wentzell, S.S. Anna-Arriola, A.L. Rodriguez, A.D. Aragon, G.A. Quiñones, C. Allen, and M. Werner-Washburne, "Genomic Analysis of Stationary-Phase and Exit inSaccharomyces cerevisiae: Gene Expression and Identification of Novel Essential Genes", Molecular Biology of the Cell, vol. 15, pp. 5295-5305, 2004. http://dx.doi.org/10.1091/mbc.e03-11-0856
- L. Galdieri, S. Mehrotra, S. Yu, and A. Vancura, "Transcriptional Regulation in Yeast during Diauxic Shift and Stationary Phase", OMICS: A Journal of Integrative Biology, vol. 14, pp. 629-638, 2010. http://dx.doi.org/10.1089/omi.2010.0069
- J.J.M. Ter Linde, and H.Y. Steensma, "A microarray‐assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae", Yeast, vol. 19, pp. 825-840, 2002. http://dx.doi.org/10.1002/yea.879
- C.T. Harbison, D.B. Gordon, T.I. Lee, N.J. Rinaldi, K.D. Macisaac, T.W. Danford, N.M. Hannett, J. Tagne, D.B. Reynolds, J. Yoo, E.G. Jennings, J. Zeitlinger, D.K. Pokholok, M. Kellis, P.A. Rolfe, K.T. Takusagawa, E.S. Lander, D.K. Gifford, E. Fraenkel, and R.A. Young, "Transcriptional regulatory code of a eukaryotic genome", Nature, vol. 431, pp. 99-104, 2004. http://dx.doi.org/10.1038/nature02800
- T. Keng, "HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae.", Molecular and Cellular Biology, vol. 12, pp. 2616-2623, 1992. http://dx.doi.org/10.1128/MCB.12.6.2616
- A.P. Gasch, P.T. Spellman, C.M. Kao, O. Carmel-Harel, M.B. Eisen, G. Storz, D. Botstein, and P.O. Brown, "Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes", Molecular Biology of the Cell, vol. 11, pp. 4241-4257, 2000. http://dx.doi.org/10.1091/mbc.11.12.4241
- N. Rachidi, M. Martinez, P. Barre, and B. Blondin, "Saccharomyces cerevisiae PAU genes are induced by anaerobiosis", Molecular Microbiology, vol. 35, pp. 1421-1430, 2000. http://dx.doi.org/10.1046/j.1365-2958.2000.01807.x
- C. VERDUYN, E. POSTMA, W.A. SCHEFFERS, and J.P. VAN DIJKEN, "Energetics of Saccharomyces Cerevisiae in Anaerobic Glucose-Limited Chemostat Cultures", Journal of General Microbiology, vol. 136, pp. 405-412, 1990. http://dx.doi.org/10.1099/00221287-136-3-405
- D.E. Atkinson, "Energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers", Biochemistry, vol. 7, pp. 4030-4034, 1968. http://dx.doi.org/10.1021/bi00851a033
- C. Verduyn, E. Postma, W.A. Scheffers, and J.P. van Dijken, "Physiology of Saccharomyces Cerevisiae in Anaerobic Glucose-Limited Chemostat Culturesx", Journal of General Microbiology, vol. 136, pp. 395-403, 1990. http://dx.doi.org/10.1099/00221287-136-3-395
- J. Choi, and C. Lee, "Maintenance of cellular ATP level by caloric restriction correlates chronological survival of budding yeast", Biochemical and Biophysical Research Communications, vol. 439, pp. 126-131, 2013. http://dx.doi.org/10.1016/j.bbrc.2013.08.014
- W.J. Ball, and D.E. Atkinson, "Adenylate energy charge in Saccharomyces cerevisiae during starvation.", Journal of bacteriology, 1975. http://www.ncbi.nlm.nih.gov/pubmed/1090610
- B. Sonnleitner, and O. Käppeli, "Growth of Saccharomyces cerevisiae is controlled by its limited respiratory capacity: Formulation and verification of a hypothesis", Biotechnology and Bioengineering, vol. 28, pp. 927-937, 1986. http://dx.doi.org/10.1002/bit.260280620
- E. Thomsson, C. Larsson, E. Albers, A. Nilsson, C.J. Franzén, and L. Gustafsson, "Carbon Starvation Can Induce Energy Deprivation and Loss of Fermentative Capacity in Saccharomyces cerevisiae", Applied and Environmental Microbiology, vol. 69, pp. 3251-3257, 2003. http://dx.doi.org/10.1128/AEM.69.6.3251-3257.2003
- G.G. Perrone, S. Tan, and I.W. Dawes, "Reactive oxygen species and yeast apoptosis", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1354-1368, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.01.023
- C. Lu, M.J. Brauer, and D. Botstein, "Slow Growth Induces Heat-Shock Resistance in Normal and Respiratory-deficient Yeast", Molecular Biology of the Cell, vol. 20, pp. 891-903, 2009. http://dx.doi.org/10.1091/mbc.E08-08-0852
- L.G.M. Boender, E.A.F. de Hulster, A.J.A. van Maris, P.A.S. Daran-Lapujade, and J.T. Pronk, "Quantitative Physiology of Saccharomyces cerevisiae at Near-Zero Specific Growth Rates", Applied and Environmental Microbiology, vol. 75, pp. 7578-7578, 2009. http://dx.doi.org/10.1128/aem.02344-09
- L.G. Boender, A.J. Maris, E.A. Hulster, M.J. Almering, I.J. Klei, M. Veenhuis, J.H. Winde, J.T. Pronk, and P. Daran-Lapujade, "Cellular responses of Saccharomyces cerevisiae at near-zero growth rates: transcriptome analysis of anaerobic retentostat cultures", FEMS Yeast Research, vol. 11, pp. 603-620, 2011. http://dx.doi.org/10.1111/j.1567-1364.2011.00750.x
- N.A. Binai, M.M.M. Bisschops, B. van Breukelen, S. Mohammed, L. Loeff, J.T. Pronk, A.J.R. Heck, P. Daran-Lapujade, and M. Slijper, "Proteome Adaptation of Saccharomyces cerevisiae to Severe Calorie Restriction in Retentostat Cultures", Journal of Proteome Research, vol. 13, pp. 3542-3553, 2014. http://dx.doi.org/10.1021/pr5003388
- E. Swinnen, V. Wanke, J. Roosen, B. Smets, F. Dubouloz, I. Pedruzzi, E. Cameroni, C. De Virgilio, and J. Winderickx, "Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae", Cell Division, vol. 1, 2006. http://dx.doi.org/10.1186/1747-1028-1-3
- M. Wei, P. Fabrizio, J. Hu, H. Ge, C. Cheng, L. Li, and V.D. Longo, "Life Span Extension by Calorie Restriction Depends on Rim15 and Transcription Factors Downstream of Ras/PKA, Tor, and Sch9", PLoS Genetics, vol. 4, pp. e13, 2008. http://dx.doi.org/10.1371/journal.pgen.0040013
- M.M. Bisschops, P. Zwartjens, S.G. Keuter, J.T. Pronk, and P. Daran-Lapujade, "To divide or not to divide: A key role of Rim15 in calorie-restricted yeast cultures", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1843, pp. 1020-1030, 2014. http://dx.doi.org/10.1016/j.bbamcr.2014.01.026
- E. Thomsson, M. Svensson, and C. Larsson, "Rapamycin pre-treatment preserves viability, ATP level and catabolic capacity during carbon starvation ofSaccharomyces cerevisiae", Yeast, vol. 22, pp. 615-623, 2005. http://dx.doi.org/10.1002/yea.1219
- P.A. Gibney, C. Lu, A.A. Caudy, D.C. Hess, and D. Botstein, "Yeast metabolic and signaling genes are required for heat-shock survival and have little overlap with the heat-induced genes", Proceedings of the National Academy of Sciences, vol. 110, 2013. http://dx.doi.org/10.1073/pnas.1318100110
- S.D. Westerheide, and R.I. Morimoto, "Heat Shock Response Modulators as Therapeutic Tools for Diseases of Protein Conformation", Journal of Biological Chemistry, vol. 280, pp. 33097-33100, 2005. http://dx.doi.org/10.1074/jbc.R500010200
- S. Lindquist, and G. Kim, "Heat-shock protein 104 expression is sufficient for thermotolerance in yeast.", Proceedings of the National Academy of Sciences, vol. 93, pp. 5301-5306, 1996. http://dx.doi.org/10.1073/pnas.93.11.5301
- M. Haslbeck, "Hsp26: a temperature-regulated chaperone", The EMBO Journal, vol. 18, pp. 6744-6751, 1999. http://dx.doi.org/10.1093/emboj/18.23.6744
- U.M. Praekelt, and P.A. Meacock, "HSP12, a new small heat shock gene of Saccharomyces cerevisiae: Analysis of structure, regulation and function", Molecular and General Genetics MGG, vol. 223, pp. 97-106, 1990. http://dx.doi.org/10.1007/bf00315801
- M. Werner-Washburne, D.E. Stone, and E.A. Craig, "Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae.", Molecular and Cellular Biology, vol. 7, pp. 2568-2577, 1987. http://dx.doi.org/10.1128/MCB.7.7.2568
- E.J. Dufourc, "Sterols and membrane dynamics", Journal of Chemical Biology, vol. 1, pp. 63-77, 2008. http://dx.doi.org/10.1007/s12154-008-0010-6
-
S. Guyot, P. Gervais, M. Young, P. Winckler, J. Dumont, and H.M. Davey, "Surviving the heat: heterogeneity of response in
S accharomyces cerevisiae provides insight into thermal damage to the membrane", Environmental Microbiology, vol. 17, pp. 2982-2992, 2015. http://dx.doi.org/10.1111/1462-2920.12866 - L. Caspeta, Y. Chen, P. Ghiaci, A. Feizi, S. Buskov, B.M. Hallström, D. Petranovic, and J. Nielsen, "Altered sterol composition renders yeast thermotolerant", Science, vol. 346, pp. 75-78, 2014. http://dx.doi.org/10.1126/science.1258137
- V.E. CHESTER, "The dissimilation of the carbohydrate reserves of a strain of Saccharomyces cerevisiae.", The Biochemical journal, 1963. http://www.ncbi.nlm.nih.gov/pubmed/14020679
- C.W.T. van Roermund, H.R. Waterham, L. Ijlst, and R.J.A. Wanders, "Fatty acid metabolism in Saccharomyces cerevisiae", Cellular and Molecular Life Sciences (CMLS), vol. 60, pp. 1838-1851, 2003. http://dx.doi.org/10.1007/s00018-003-3076-x
- C. Verduyn, A.H. Stouthamer, W.A. Scheffers, and J.P. van Dijken, "A theoretical evaluation of growth yields of yeasts", Antonie van Leeuwenhoek, vol. 59, pp. 49-63, 1991. http://dx.doi.org/10.1007/BF00582119
- A.H. Stouthamer, "A theoretical study on the amount of ATP required for synthesis of microbial cell material", Antonie van Leeuwenhoek, vol. 39, pp. 545-565, 1973. http://dx.doi.org/10.1007/BF02578899
- N. Ziv, N.J. Brandt, and D. Gresham, "The Use of Chemostats in Microbial Systems Biology", Journal of Visualized Experiments, 2013. http://dx.doi.org/10.3791/50168
- J. Büchs, "Introduction to advantages and problems of shaken cultures", Biochemical Engineering Journal, vol. 7, pp. 91-98, 2001. http://dx.doi.org/10.1016/s1369-703x(00)00106-6
- S. Nagarajan, A.L. Kruckeberg, K.H. Schmidt, E. Kroll, M. Hamilton, K. McInnerney, R. Summers, T. Taylor, and F. Rosenzweig, "Uncoupling reproduction from metabolism extends chronological lifespan in yeast", Proceedings of the National Academy of Sciences, vol. 111, 2014. http://dx.doi.org/10.1073/pnas.1323918111
- S.S. Lee, I.A. Vizcarra, D.H.E.W. Huberts, L.P. Lee, and M. Heinemann, "Whole lifespan microscopic observation of budding yeast aging through a microfluidic dissection platform", Proceedings of the National Academy of Sciences, vol. 109, pp. 4916-4920, 2012. http://dx.doi.org/10.1073/pnas.1113505109
- L. Basso, T. Basso, and S. Rocha, "Ethanol production in Brazil: the industrial process and its impact on yeast fermentation", In: dos Santos Bernardes MA, editor. Biofuel production - Recent developments and prospects. Intech, Rijeka; pp 85-100., 2011.
- B.R. Gibson, S.J. Lawrence, J.P.R. Leclaire, C.D. Powell, and K.A. Smart, "Yeast responses to stresses associated with industrial brewery handling: Figure 1", FEMS Microbiology Reviews, vol. 31, pp. 535-569, 2007. http://dx.doi.org/10.1111/j.1574-6976.2007.00076.x
- A. Hagman, and J. Piškur, "A Study on the Fundamental Mechanism and the Evolutionary Driving Forces behind Aerobic Fermentation in Yeast", PLOS ONE, vol. 10, pp. e0116942, 2015. http://dx.doi.org/10.1371/journal.pone.0116942
- M.R. Goddard, and D. Greig, "Saccharomyces cerevisiae: a nomadic yeast with no niche?", FEMS Yeast Research, vol. 15, 2015. http://dx.doi.org/10.1093/femsyr/fov009
- E. Reifenberger, E. Boles, and M. Ciriacy, "Kinetic Characterization of Individual Hexose Transporters of Saccharomyces Cerevisiae and their Relation to the Triggering Mechanisms of Glucose Repression", European Journal of Biochemistry, vol. 245, pp. 324-333, 1997. http://dx.doi.org/10.1111/j.1432-1033.1997.00324.x
- K. Entian, and P. Kötter, "25 Yeast Genetic Strain and Plasmid Collections", Methods in Microbiology, pp. 629-666, 2007. http://dx.doi.org/10.1016/s0580-9517(06)36025-4
- J.F. Nijkamp, M. van den Broek, E. Datema, S. de Kok, L. Bosman, M.A. Luttik, P. Daran-Lapujade, W. Vongsangnak, J. Nielsen, W.H. Heijne, P. Klaassen, C.J. Paddon, D. Platt, P. Kötter, R.C. van Ham, M.J. Reinders, J.T. Pronk, D. de Ridder, and J. Daran, "De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology", Microbial Cell Factories, vol. 11, 2012. http://dx.doi.org/10.1186/1475-2859-11-36
- C. Verduyn, E. Postma, W.A. Scheffers, and J.P. Van Dijken, "Effect of benzoic acid on metabolic fluxes in yeasts: A continuous‐culture study on the regulation of respiration and alcoholic fermentation", Yeast, vol. 8, pp. 501-517, 1992. http://dx.doi.org/10.1002/yea.320080703
- E. Postma, C. Verduyn, W.A. Scheffers, and J.P. Van Dijken, "Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae.", Applied and environmental microbiology, 1989. http://www.ncbi.nlm.nih.gov/pubmed/2566299
- A.B. Canelas, C. Ras, A. ten Pierick, J.C. van Dam, J.J. Heijnen, and W.M. van Gulik, "Leakage-free rapid quenching technique for yeast metabolomics", Metabolomics, vol. 4, pp. 226-239, 2008. http://dx.doi.org/10.1007/s11306-008-0116-4
- J.L. Parrou, and J. François, "A Simplified Procedure for a Rapid and Reliable Assay of both Glycogen and Trehalose in Whole Yeast Cells", Analytical Biochemistry, vol. 248, pp. 186-188, 1997. http://dx.doi.org/10.1006/abio.1997.2138
- K. Johnson, R. Kleerebezem, and M.C. van Loosdrecht, "Model‐based data evaluation of polyhydroxybutyrate producing mixed microbial cultures in aerobic sequencing batch and fed‐batch reactors", Biotechnology and Bioengineering, vol. 104, pp. 50-67, 2009. http://dx.doi.org/10.1002/bit.22380
- D. Jaworek, and J. Welsch, "Adenosine 5'-diphosphate and adenosine 5'-monophosphate", In: Bergmeyer HU, Bergmeyer J, Gral M, editors. Methods of enzymatic analysis. Verlagsgesellschaft mbH, Weinheim; pp 365-370., 1985.
- M.D. Piper, P. Daran-Lapujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsen, and J.T. Pronk, "Reproducibility of Oligonucleotide Microarray Transcriptome Analyses", Journal of Biological Chemistry, vol. 277, pp. 37001-37008, 2002. http://dx.doi.org/10.1074/jbc.m204490200
- F. Mendes, S. Sieuwerts, E. de Hulster, M.J.H. Almering, M.A.H. Luttik, J.T. Pronk, E.J. Smid, P.A. Bron, and P. Daran-Lapujade, "Transcriptome-Based Characterization of Interactions between Saccharomyces cerevisiae and Lactobacillus delbrueckii subsp. bulgaricus in Lactose-Grown Chemostat Cocultures", Applied and Environmental Microbiology, vol. 79, pp. 5949-5961, 2013. http://dx.doi.org/10.1128/AEM.01115-13
- M. Hebly, D. de Ridder, E.A.F. de Hulster, P. de la Torre Cortes, J.T. Pronk, and P. Daran-Lapujade, "Physiological and Transcriptional Responses of Anaerobic Chemostat Cultures of Saccharomyces cerevisiae Subjected to Diurnal Temperature Cycles", Applied and Environmental Microbiology, vol. 80, pp. 4433-4449, 2014. http://dx.doi.org/10.1128/AEM.00785-14
- P. Daran-Lapujade, M.L. Jansen, J. Daran, W. van Gulik, J.H. de Winde, and J.T. Pronk, "Role of Transcriptional Regulation in Controlling Fluxes in Central Carbon Metabolism of Saccharomyces cerevisiae", Journal of Biological Chemistry, vol. 279, pp. 9125-9138, 2004. http://dx.doi.org/10.1074/jbc.m309578200
- M. Teste, M. Duquenne, J.M. François, and J. Parrou, "Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae", BMC Molecular Biology, vol. 10, 2009. http://dx.doi.org/10.1186/1471-2199-10-99
- T.A. Knijnenburg, J.H. de Winde, J. Daran, P. Daran-Lapujade, J.T. Pronk, M.J. Reinders, and L.F. Wessels, "Exploiting combinatorial cultivation conditions to infer transcriptional regulation", BMC Genomics, vol. 8, 2007. http://dx.doi.org/10.1186/1471-2164-8-25
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We thank for Pauline Folch, Nuria Barrajón-Simanca, Xavier Hakkaart for assistance in performing the experiments. Martínez-Moreno, R is recipient of a CSIC JAE-Predoc grant co-funded by the European Social Fund of the EU.
COPYRIGHT
© 2015

Oxygen availability strongly affects chronological lifespan and thermotolerance in batch cultures of Saccharomyces cerevisiae by Markus M.M. Bisschops et al. is licensed under a Creative Commons Attribution 4.0 International License.