Research Articles:
Microbial Cell, Vol. 3, No. 11, pp. 540 - 553; doi: 10.15698/mic2016.11.538
The ubiquitin-conjugating enzyme, Ubc1, indirectly regulates SNF1 kinase activity via Forkhead-dependent transcription
1 Department of Anatomy and Cell Biology, University of Saskatchewan, Saskatoon, SK, Canada S7N 5E5.
2 Department of Medicine, University of Saskatchewan, Saskatoon, SK, Canada S7N 5E5.
Keywords: SNF1 kinase, Ubc1, Forkheads, APC, protein stability.
Abbreviations:
AMPK - AMP-dependent protein kinases,
APC - anaphase promoting complex,
CM - complete medium,
ERAD - endoplasmic reticulum associated degradation,
FACS - fluorescent activated cell sorting,
Fkh1/2 - Forkhead 1/2,
GFP – green fluorescent protein,
Ub – Ubiquitin,
UBA - Ubiquitin-associated domain.
Received originally: 03/06/2016 Received in revised form: 16/10/2016
Accepted: 21/10/2016
Published: 05/11/2016
Correspondence:
Terra G. Arnason, terra.arnason@usask.ca
Conflict of interest statement: The authors declare no competing interests.
Please cite this article as: Rubin Jiao, Liubov Lobanova, Amanda Waldner, Anthony Fu, Linda Xiao, Troy A. Harkness, and Terra G. Arnason (2016). The Ubiquitin-Conjugating Enzyme, Ubc1, Indirectly Regulates SNF1 Kinase Activity Via Forkhead-dependent Transcription. Microbial Cell 3(11): 540-553.
Abstract
The SNF1 kinase in Saccharomyces cerevisiae is an excellent model to study the regulation and function of the AMP-dependent protein kinase (AMPK) family of serine-threonine protein kinases. Yeast discoveries regarding the regulation of this non-hormonal sensor of metabolic/environmental stress are conserved in higher eukaryotes, including poly-ubiquitination of the α-subunit of yeast (Snf1) and human (AMPKα) that ultimately effects subunit stability and enzyme activity. The ubiquitin-cascade enzymes responsible for targeting Snf1 remain unknown, leading us to screen for those that impact SNF1 kinase function. We identified the E2, Ubc1, as a regulator of SNF1 kinase function. The decreased Snf1 abundance found upon deletion of Ubc1 is not due to increased degradation, but instead is partly due to impaired SNF1 gene expression, arising from diminished abundance of the Forkhead 1/2 proteins, previously shown to contribute to SNF1 transcription. Ultimately, we report that the Fkh1/2 cognate transcription factor, Hcm1, fails to enter the nucleus in the absence of Ubc1. This implies that Ubc1 acts indirectly through transcriptional effects to modulate SNF1 kinase activity.
INTRODUCTION
The SNF1 kinase class of serine/threonine kinases, which includes the AMP-dependent protein kinase (AMPK) in other systems, are of widespread interest because of their important roles in glucose homeostasis, stress resistance, and aging [1][2][3]. These enzymes are inactive under optimal conditions, yet are rapidly activated in response to a wide variety of nutritional and stress cues. The active kinases, in turn, exert their activity to alter cellular pathways at the protein and transcriptional level to maintain homeostasis or to direct adaptive mechanisms for stress resistance. Simply, low glucose growth conditions will activate SNF1 kinase in yeast, whereas muscle contraction or fasting will do the same in animals [3][4][5]. The dominant, essential, and finely responsive regulatory step is the phosphorylation of Snf1 by upstream kinases, balanced by its controlled dephosphorylation [4][6]. This governing event is conserved between yeast and all higher eukaryotes. In yeast, SNF1 kinase directly phosphorylates a variety of downstream targets, including the nuclear target Mig1 [7] and the cytosolic target Rod1 [8]. Mig1 functions as a transcriptional inhibitor that prevents SUC2 expression, whereas Rod1 is involved in glucose transporter endocytosis [8]. Several other described events act on SNF1 kinase including allosteric tightening of the Snf1 (α) and Snf4 (γ) subunits within the heterotrimeric complex (by convention, note lower case for the yeast protein α subunit [Snf1], upper case for the enzyme complex [SNF1 kinase]) [9], and the nuclear accumulation of the Snf1-Gal83 (β subunit)-Snf4 enzyme within the nucleus, necessary for transcriptional changes [10]. In addition, our recent work reported that the yeast orthologs of mammalian Forkhead Box (Fox) transcription factors, Forkhead (Fkh) 1 and 2, regulate SNF1 gene transcription, impacting the protein abundance of the Snf1 subunit [11]. The Snf1 subunit contains autoinhibitory and ubiquitin-associated (UBA) domains that act as restraining functions on activity [11][12]. More recently, additional negative influences have been shown to occur through Snf1 subunit posttranslational modifications, specifically ubiquitination and SUMOylation, which effectively decreased Snf1 protein abundance through degradation, with resulting reductions in SNF1 kinase activity [13][14][15]. Ubiquitin (Ub) becomes covalently attached to target proteins through the sequential action of Ub activating (E1), conjugating (E2), and ligase (E3) activities: in yeast there is a single E1, a finite well-described group of eleven E2s, and an ever-expanding recognition of E3 ligase activities.
–
Our goal was to identify discrete E2s that are involved in SNF1 kinase activity in response to glucose levels and anticipated revealing those which are involved in Snf1-Ub attachment. Here, we report that the cell cycle and stress-related E2, Ubc1, indirectly affects SNF1 kinase activity not through stability, but through upstream events effecting the yeast Fox orthologs Fkh1/2 that provide transcriptional control of the Snf1 subunit. Ubc1 is known to act, along with Ubc4, with the anaphase promoting complex (APC) to target and polyubiquitinate APC substrates for cell cycle related degradation to enable exit from metaphase, and entry to G1 [15]. Intriguingly, the ubc1∆ mitotic arrest point is consistent with a disruption of APC-dependent exit from mitosis and failure to enter G1 [16]. Nonetheless, our data suggest that Ubc1 acts on Hcm1 in an APC-independent manner. Our observations suggest a model where Hcm1 is modified in a Ubc1- dependent manner to facilitate Hcm1 nuclear shuttling and activation of the Fkh/Snf1 stress response pathway.
RESULTS
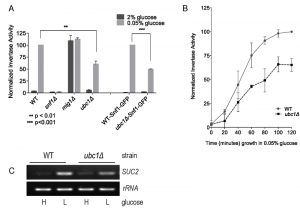
Deletion of the E2 enzyme, Ubc1, impairs SNF1 kinase-dependent invertase activity.
When glucose is limiting, yeast adapts to using alternative carbon sources. A key function of the SNF1 kinase is to adapt metabolic pathways to non-glucose carbohydrate sources, and the mechanism is particularly well documented for sucrose. Sucrose utilization requires the expression of invertase, an enzyme that cleaves the disaccharide sucrose molecule into glucose and fructose, which is encoded by the SUC2 gene [17]. When glucose is abundant, SUC2 expression is repressed by the binding of the transcriptional repressor, Mig1 to the SUC2 promotor. Under limiting glucose conditions, activated SNF1 kinase enters the nucleus, phosphorylates Mig1 protein via its inherent kinase activity and releases Mig1 from the SUC2 promoter. The subsequent expression of SUC2 can be quantitatively determined by colorimetric “Invertase assay” [18] or directly through RT-PCR, as an indirect measure of SNF1 kinase activity.
–
Fig. 1A demonstrates the expected rise in invertase activity after 2 hours of growth in low (0.05%) glucose in a wild type (WT) yeast strain, and the complete dependence of this on the Snf1 α catalytic subunit (snf1Δ). In contrast, disruption of the SUC2 transcriptional repressor Mig1 (mig1Δ) [7] demonstrates high activity regardless of glucose levels, as expected for loss of regulated repression. Disruption of the UBC1 gene (ubc1Δ) resulted in a significant decrease in invertase activity as compared to WT. This defect was also observed in WT and ubc1Δ strains carrying the endogenous green-fluorescent (GFP) protein Snf1-GFP fusion into either strain (WT-Snf1-GFP or ubc1Δ-Snf1-GFP). To expand on this, we also tested invertase activity at short intervals leading up to a 2 hour time point (Fig. 1B) and report the presence of early (1 hour) and sustained impairment of invertase activity. The corresponding changes to SUC2 expression in the WT and ubc1Δ strains under activating and repressive conditions paralleled that of the invertase RNA (Fig. 1C).
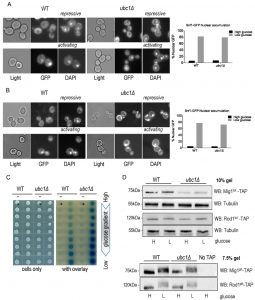
Loss of Ubc1 function does not impair SNF1 kinase nuclear accumulation, allosteric associations or substrate targeting.
Invertase assay defects (or decreases in maximal SUC2 expression) can arise from disruption of any of multiple steps in SNF1 kinase activation, including protein abundance, activating phosphorylation, allosteric associations, nuclear import, or phosphorylation of Mig1. These stages can be isolated and independently assessed to pinpoint where Ubc1 affects SNF1 kinase activity. First, we asked if Ubc1 was required for movement of the Snf1-Gal83-Snf4 kinase complex into the nucleus under activating conditions, as a failure to efficiently accumulate in the nucleus would explain the impairment in transcriptional release of SUC2 expression. We initially expressed Snf1-GFP constitutively from a 2μ yeast plasmid transformed into WT and ubc1Δ strains and used live fluorescent microscopy of these isogenic strains to localize Snf1-GFP to the nucleus (identified by DAPI staining) (Fig. 2A). The plasmid-expressed Snf1 subunit rapidly relocates to the nucleus after stimulating conditions in a manner indistinguishable from WT [10]. Furthermore, endogenous expression of a genomic version of GFP-tagged Snf1 subunit did not alter the efficiency of nuclear accumulation in WT or ubc1Δ isogenic strains (Fig. 2B). Multiple biological repeats of these experiments allowed us to score the relative efficiency of nuclear import of Snf1-GFP in these WT and ubc1Δ strains, demonstrating that there is no impairment of nuclear accumulation in the absence of the functional Ubc1 protein.
Next, we assessed if the UBC1 disruption interfered with the allosteric associations between the α and γ subunits of the kinase upon activation. 2-hybrid analysis was used to compare Snf1-Snf4 interactions throughout a glucose gradient; β-galactosidase production results in a visible blue color and correlates with the strength of the associations between these two proteins [11]. In WT yeast, we observed increased Snf1-Snf4 associations within the 2-hybrid assay as the glucose concentration drops, which is also seen in the isogenic ubc1Δ strain, at even greater levels than WT (Fig. 2C). Clearly, the invertase defect linked with UBC1 disruption is not related to allosteric hindrance.
–
We next asked if the UBC1 deletion affected the ability of SNF1 kinase to phosphorylate known protein targets. De-repression of SUC2 requires SNF1 kinase-dependent phosphorylation of the nuclear transcriptional repressor protein, Mig1. Non-nuclear targets for SNF1 kinase phosphorylation include the Rod1 protein that resides at the plasma membrane [8]. Glucose responsive phosphorylation of both nuclear Mig1 and cytosolic Rod1 can be directly assessed by Western analysis by their visible phospho-shift to higher molecular weights [7][8]. We find a noticeable upwards phospho-shift of each target under low glucose in both WT and ubc1Δ strains (Fig. 2D). Although there is an apparent decrease in Mig1 protein abundance in the ubc1Δ strain, the phospho-shift is not impaired, nor is there a defect in the expected Mig1 nuclear export under activating conditions (Fig. S1). Therefore, Ubc1 deletion does not impair the enzymatic activity of SNF1 kinase.
–
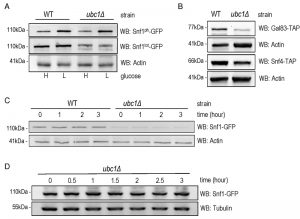
Snf1 protein abundance, but neither stability nor phosphorylation, is decreased by Ubc1 disruption.
An obvious question to ask was whether the role of Ubc1 in SNF1 kinase regulation was to target Snf1 for ubiquitination, and ultimately degradation. It is known that the catalytic Snf1 α-subunit can be polyubiquitinated and thereby effect its abundance [14]. We compared the steady-state abundance of endogenous Snf1-GFP protein in logarithmically growing WT and ubc1Δ strains (Fig. 3A) and observed a clear decrease in Snf1 abundance, irrespective of the activation state of the enzyme, limited to the ubc1Δ strain. This is not consistent with Ubc1-dependent ubiquitination and subsequent degradation of Snf1, as this would instead manifest as an increase in Snf1 protein in the UBC1 deletion. Activating Snf1 phosphorylation was maintained at near-WT levels despite the decrease in total Snf1 protein (Fig. 3A). In addition to Snf1, protein levels of both the endogenous Snf4 γ- and Gal83 β-subunits were down in the ubc1Δ strain, compared to WT (Fig. 3B). To directly assess if there was enhanced degradation of Snf1 in the absence of Ubc1 function, we performed cycloheximide protein degradation assays of Snf1 over a 3-hour period (Fig. 3C) in WT and ubc1Δ strains. Snf1 was stable over the three-hour period in the WT strain and also appeared stable in ubc1Δ. To confirm the stability of Snf1 in the ubc1Δ strain, a biological repeat was performed with a greater protein load (80 μg/lane versus 40 μg/lane) (Fig. 3D). These results suggest that Ubc1 does not play a role in Snf1 protein stability.
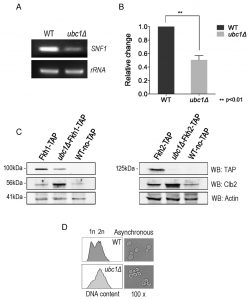
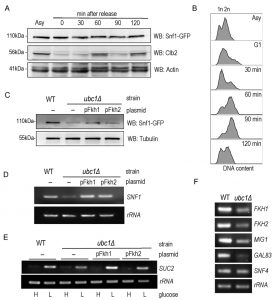
SNF1 expression and Fkh protein abundance are decreased upon Ubc1 deletion.
We addressed the possibility that SNF1 transcription was decreased in the ubc1Δ strain, as an explanation for the decreased abundance of the Snf1 protein. RT-PCR of the SNF1 gene revealed a clear decrease in transcription in the ubc1Δ strain (Fig. 4A), being approximately 50% that of the isogenic WT (Fig. 4B). We had previously found the yeast Fox orthologs, Fkh 1 and Fkh2, to be involved in SNF1 gene expression [11] and asked if the absence of Ubc1 function was affecting Fkh1/2 activity, upstream of Snf1. In asynchronous cells, we observed that disruption of UBC1 noticeably diminished Fkh1 protein abundance, while Fkh2 protein was essentially absent in the same strain even when significantly more protein (lysate) was tested (80 μg/lane for Fkh2 versus 15 μg/lane for Fkh1) (Fig. 4C). Fkh1 and Fkh2 are known to be transcriptionally regulated in synchrony with the cell cycle under the influence of a third forkhead member, Hcm1 [19]. Yeast strains deleted for UBC1 have been found to be associated with cell cycle defects and to accumulate with large buds in late G2/M phase [16]. Indeed, we found that the protein abundance of endogenous Clb2 was markedly elevated in the ubc1Δ strain as compared to WT, consistent with an accumulation of cells residing in G2/M phase when Clb2 levels are highest [20]. To confirm this, flow cytometry of early logarithmic (OD600 of 0.4) asynchronous yeast cells from ubc1Δ strains demonstrated a significant inherent accumulation of cells with fully replicated DNA (2n) in G2/M (Fig. 4D). Light microscopy of cells representative of those undergoing flow cytometry show a heterogeneous population in WT with various bud sizes, yet a clear accumulation of large budded yeast cells in ubc1Δ, consistent with previous reports of G2/M arrest [16]. The published report noted that FKH1 and FKH2 transcript levels are highest in G2/M phase and lowest in G1 [19] was inconsistent with the decreased Forkhead protein levels we observed in the ubc1Δ strain partially stalled at G2/M, leading us to investigate this further.
Snf1 protein, and glucose-responsive SUC2 expression, levels are nearly reestablished by Fkh1 or Fkh2 overexpression in the ubc1Δ strain.
Given that Fkh1 and Fkh2 can drive the expression of SNF1 [11], and that Fkh1/2 gene expression fluctuates with the cell cycle, we asked if Snf1 protein levels fluctuate in synchrony with the cell cycle. There have been no reports of SNF1 kinase being regulated in a cell cycle dependent manner to the best of our knowledge. A G1 arrest-release experiment was performed in WT yeast to determine the inherent abundance of the Snf1 subunit protein level throughout the cell cycle. Fig. 5A reveals that Snf1 protein levels do not change as synchronized cells exit G1 and pass through the cell cycle, with fluctuations of Clb2 used as a surrogate marker for passage through the cell cycle [21]. Further confirmation of successful synchronization comes from the flow cytometry analysis of these cells, showing the gradual shift of 1n to 2n (replicated) DNA (Fig. 5B). To ascertain if the decreased Snf1 abundance was a simple result of limited forkhead proteins, Fkh1 and Fkh2 were constitutively expressed in the ubc1Δ strain. The resulting Snf1 protein (Fig. 5C) and SNF1 transcripts (Fig. 5D) are increased with Fkh1, which is not as apparent with Fkh2. Advancing one step further, we similarly investigated if Fkh1 or Fkh2 expression in the ubc1Δ strain restored the low-glucose-activated SUC2 expression to that of WT levels. Fig. 5E shows that low glucose-induced expression of SUC2 is at, or higher than, WT levels in the presence of the Fkh1 plasmid. To distinguish between etiologies underlying the decreased protein abundance of Fkh1 and Fkh2 (Fig. 4C), Snf4 and Gal83 (Fig. 3B) and Mig1 (Fig. 2D), we asked if the ubc1Δ deletion also affected their transcription. With the exception of Snf4, all RT-PCR products were of lower abundance in the ubc1Δ strain (Fig. 5F). Our data suggest that Fkh1/2 transcription of SNF1 does not fully control Snf1 protein abundance in the ubc1Δ strain; despite compensated SNF1 mRNA levels (Fig. 5D), Snf1 protein levels did not reach that of wildtype (Fig. 5C). Furthermore, constitutive plasmid expression of Snf1-HA within the ubc1Δ strain did not result in WT levels of Snf1 protein (Fig. S2A), suggesting that Snf1-HA is decreased in the ubc1Δ strain regardless of its endogenous transcription. Finally, we tested whether over-expression of the Snf4 γ-subunit would enhance Snf1 proteins levels, and concluded that it did not (Fig. S2B).
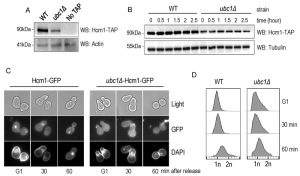
Hcm1 is impaired in its cell cycle-dependent nuclear import, upon deletion of Ubc1.
We sought a more detailed explanation for the decrease in Fkh1 and Fhk2 expression within the ubc1Δ strain, and focused on Hcm1, the forkhead family member known to regulate Fkh1 and Fkh2 expression in a cell cycle dependent manner [19]. We tested both Hcm1 protein abundance, and its ability to shuttle between the cytosol and nucleus. We first found that the steady-state protein abundance of Hcm1 was modestly decreased upon disruption of the UBC1 gene (Fig. 6A), although the protein appeared stable over an extended three-hour period (Fig. 6B). Hcm1 exhibits regulated nuclear import during G1 of the cell cycle, and we asked if the expected nuclear accumulation of Hcm1 in G1 [22] is, in fact, disrupted by deletion of the UBC1 gene. Fluorescent microscopy was used to determine endogenous Hcm1-GFP location within cells arrested in G1 and then released. We observed a clear nuclear accumulation in arrested WT cells, in direct contrast to a lack of nuclear-GFP signal in the ubc1Δ strain arrested in G1 (Fig. 6C). The morphology of the yeast cells, combined with the flow analysis (Fig. 6D) confirms that G1 arrest was successful in wildtype and ubc1Δ strains.
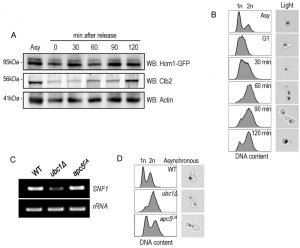
To further analyze Hcm1 protein abundance differences between the WT and ubc1Δ strains, we compared the Hcm1 protein level throughout the cell cycle after G1 arrest and release in WT. Figure 7A demonstrated the cell-cycle fluctuations in Hcm1 protein levels, fully consistent with Hcm1 levels reported by others [23]. 90 minutes after release, Hcm1 protein level reached maximum, followed by a decline at 120 minutes at which time Clb2 levels peaked, consistent with metaphase. Supporting this, flow cytometry analysis and microscopy images collected at the 120 minute timepoint are consistent with late mitosis/telophase; fully replicated (2n) and late mitosis (double budded, nuclei separated) (Fig. 7B). The doublet signal observed for the Hcm1-GFP Western analysis is not present in the Hcm1-TAP western blots, and is thus a non-specific artifact.
Ubc1 impacts SNF1 expression in an APC-independent manner.
Ubc1 is known to act with the APC to facilitate the metaphase-to-anaphase transition, and we were interested in determining whether Ubc1’s impact on transcription was APC-dependent, or –independent. We made use of yeast cells harboring the Apc5 subunit mutation (apc5CA), to test this. Figure 7C indicates that SNF1 gene expression was not affected in the apc5CA temperature sensitive strain, and was decreased only in ubc1Δ cells. We next compared if the cell cycle arrest position and cell morphology between the apc5CA and ubc1Δ strains were similar. Early logarithmic, asynchronous, yeast cells from ubc1Δ and apc5CA strains were analyzed using flow cytometry and cells were imaged using propidium iodide staining of nucleic acid (Fig. 7D). Interestingly, the inherent arrest point of the ubc1Δ strain exhibited a distinctly different 1n/2n content, and nuclear positioning to that of apc5CA cells (Fig 7D). Together, these results clearly indicate that the role of Ubc1 on SNF1 expression is independent of the APC.
–
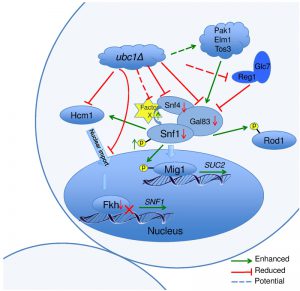
Schematic of Ubc1-dependent mechanisms and potential targets impacting SNF1 kinase activity.
Figure 8 graphically summarizes the observations made regarding the role of Ubc1 in SNF1 kinase function. In general, deletion of Ubc1 function resulted in decreased protein abundance of all three SNF1 kinase subunits tested, with transcriptional declines limited to Snf1 and Gal83 subunits, as Snf4 was not affected. The maintenance of WT-levels of Snf1 phosphorylation suggests a rebalancing of upstream kinase and phosphatase activities. The impaired SNF1 expression is due to a simultaneous decrease in FKH1 and FKH2 expression and protein levels, ultimately appearing to arise from a failure of Hcm1 to accumulate in the nucleus to facilitate their transcription. The Ubc1 protein target involved in Hcm1 shuttling is not known. Enhanced allosterism within the SNF1 kinase complex may arise from the relative hyper-phosphorylation of Snf1, or from the loss of a Factor X that contributes to steric hindrance of subunit associations.
DISCUSSION
The SNF1 kinase/AMPK family of enzymes are tightly regulated, non-hormonal, sensors of stress and nutrient availability that facilitate adaptation of cellular pathways to maintain homeostasis. The yeast SNF1 kinase is a heterotrimeric Ser/Thr protein kinase complex that is activated, in part, by energy and nutrient limitations via an essential phosphorylation step. Understanding the intricacies of the subtle regulatory mechanisms controlling their activation and activity is also of great interest. Many fields, not the least of which is the potential benefit to human health, will advance with greater knowledge of regulatory targets to enhance the activity of these enzymes.
–
Through its kinase function, activated SNF1 kinase shifts the utilization of specific metabolic pathways in order to maintain cellular ATP levels [24]. The SNF1 kinase is strongly evolutionarily conserved from yeast to humans, and fundamental mechanisms regulating SNF1 kinase activity in yeast have been proven to be likewise used in higher eukaryotes, including the essential phosphorylation on its α subunit, regulated dephosphorylation, allosteric subunit associations, and nuclear shuttling [12]. In addition to these discrete steps in SNF1 kinase activation, it has also been reported that the catalytic α subunit of the yeast SNF1 kinase, Snf1 is polyubiquitinated and degraded [14], similar to an earlier report of this mechanism acting on the human ortholog, AMPK [25]. Of note, we do not find evidence for inherent Snf1 protein instability and degradation in these experiments, irrespective of the kinase activation state or glucose availability (Fig 3A).
–
The covalent attachment of Ub to a target protein requires the concerted action of the Ub-activating (E1), Ub-conjugating (E2), and Ub-ligase (E3) activities [26], and specific protein targets are recognized and selected by specific E2/E3 combinations [16]. The identity of the E2(s) required for the reported Ub conjugation to Snf1 has not been reported in yeast or in other eukaryotes to our knowledge, and as an approach to identify these factors, we systematically screened E2 deletion strains for effects on invertase activity, a SNF1 kinase-dependent event. Given the well-known consequence of protein stability changes upon ubiquitination, we had predicted that deletion of important E2 enzymes in Snf1-Ub targeting would create a stabilizing effect on Snf1, with the possibility of enhanced SNF1 activity due to Snf1 accumulation.
–
Here, we do not report the E2 involved in targeting Snf1 for ubiquitination. Instead, we report that deletion of the gene encoding the E2, UBC1, resulted in a ~50% reduction of SNF1 kinase activity that did not arise from deficiencies in glucose-regulated nuclear import or subunit associations, or in its ability to target cytosolic or nuclear targets for phosphorylation. Rather, the explanation resides in the noticeable decrease of Snf1 protein in the ubc1Δ strain, regardless of whether the growth conditions are activating or repressive for SNF1 kinase activity (Fig. 3A). It is interesting to note that, despite lower total Snf1 protein, a conspicuous and consistent enhancement of Snf1-Snf4 interactions throughout the 2-hybrid glucose gradient (Fig. 2C) and relative degree of Snf1 phosphorylation is maintained at near WT levels (Fig. 3A), yet was not capable of complementing for SNF1 kinase invertase activity. A simultaneous decrease of the β- (Gal83) and γ-(Snf4) subunits of the SNF1 kinase heterotrimer in this strain very likely contributes to this decrease in nuclear activity. This phenomenon of Snf1 hyperphosphorylation has been noticed previously [27] and has been proposed to be a compensatory mechanism to preserve SNF1 kinase function, with our results suggesting that it may also extend to allosterism. Decreases in the level of one subunit within SNF1 kinase have also been shown to correspond to decreases in the remaining subunits [28]. Accordingly, we observed decreases in the γ subunit (Snf4) and the β subunit (Gal83) in the ubc1Δ strain (Figure 3B), perhaps as a means of preserving normal stoichiometry within the enzyme complex.
–
One of the major functions of ubiquitin conjugation is to target proteins for degradation by the proteasome [29]. In contrast to protein degradation resulting from polyubiquitination through K48 chains, monoubiquitination (monoUb) of proteins has been shown to contribute to protein stability [30]. Furthermore, ubiquitin conjugation can also have other important functions unrelated to protein degradation such as subcellular trafficking and protein associations [31]. Ubc1 is known to contribute to K48 polyUb chain formation [16], whereas in vivo reports of intrinsic monoUb activity were not found. Interestingly, we found the Snf1 subunit to be stable for hours in both the WT and ubc1Δ strains (Fig. 3C), without detection of higher molecular weight bands under these physiological conditions to suggest Snf1 is polyubiquitinated. While the stability of Snf1 in the WT strain was initially unexpected, it is clear that the previously reported effects of Snf1 instability in response to SUMOylation and ubiquitination required the forced accumulation of polyUb chains attached to Snf1. This was enabled by genetic manipulation of the yeast strains, through disruption of the gene encoding the Ub-removal function, UBP8, and the resulting polyUb proteins that are targeted for proteasomal degradation [14][15]. Otherwise Snf1 was not noticeably degraded.
–
Unexpectedly, the deletion of UBC1 resulted in a substantial decrease in SNF1 expression (Fig. 4A &B), and is likely a strong contributing cause for the low Snf1 protein levels in this strain. This correlates with a simultaneous decrease in Fkh1 and Fkh2 protein and transcript levels (Fig 4C and 5F). We had previously demonstrated that Fkh1, and to a lesser extent Fkh2, are necessary for SNF1 expression [11]. Constitutive expression of either Fkh1 or Fkh2 in the ubc1Δ strain returned SNF1 transcript levels towards WT levels; Fkh1 was able to restore Snf1 protein levels and SUC2 induction to a greater extent than Fkh2 alone (Fig. 5C & E).
–
The expression of FKH1 and FKH2 fluctuates with the cell cycle, and we asked if the inherent cell cycle defect in ubc1Δ might be affecting the low Fkh1/2 levels. The ubc1Δ strain is reported to arrest as large budded cells, which we also observed (Fig. 4D). Our flow cytometry analysis confirmed the high proportion of cells with replicated DNA indicating G2/M (Fig. 4D), and our propidium iodide staining for nuclei acid shows that the nuclei have clearly separated (Fig. 2A & B, Fig. 7E), altogether consistent with a late mitotic arrest. Lastly, the clear accumulation of Clb2 in asynchronous ubc1Δ cells is known to be a result of deficient E2 activity for the APC-dependent targeting of Clb2 for degradation, which is required for cell cycle progression through M into G1 [16], explaining the arrest point of the ubc1Δ strain. The discrete differences between the apc5CA mutant and ubc1Δ cell cycle arrest points suggest that the Ubc1 cell cycle defect may be APCCdh1 mediated. APCCdh1 controls the M/G1 transition, whereas APCCdc20 controls the metaphase/anaphase transition. General defects in APC function result in large budded cells with the nucleus aligned at the bud neck, as observed in Fig. 7D. Nonetheless, our observation indicates the cell cycle arrest in ubc1Δ cells is not a result of impaired APC activity. Furthermore, the expression of FKH1/2 is normally high in G2/M in WT cells [19], yet we note an unexpected clear decrease of FKH1/2 expression in the ubc1Δ strain that is enriched for G2/M phase cells. The answer appears to be due to Ubc1 affecting Hcm1, a third member of the Forkhead transcription factor family. Hcm1 is known to drive FKH1/2 expression [19]. A decline in Hcm1 protein abundance may explain the low Fkh1/2 protein levels in the absence of Ubc1, as Hcm1 was present at lower levels in the ubc1Δ strain, even compared to WT M-phase cells (peak expression of Hcm1) (Fig. 6A, Fig. 7A). What also became evident, however, was a specific defect in Hcm1 nuclear entry: Hcm1 is known to shuttle between the cytosol and the nucleus in a cell cycle manner, which controls its availability to the promoter regions of target genes, including FKH1/2 [32]. It is interesting to find that deletion of UBC1 severely impairs the nuclear accumulation of Hcm1 in G1 arrested cells (Fig. 6B). The role of ubiquitin in regulated nuclear import of target proteins has been well established [33][34]. Accordingly, Ubc1 may normally target Hcm1 directly (or indirectly through an associated chaperone protein) for ubiquitination to promote import of Hcm1, or alternatively target a cytosolic tether for degradation. The identity of such a putative Ubc1 target controlling Hcm1 nuclear import was not revealed in this study, and we were unsuccessful in identifying Hcm1-Ub conjugates through co-immunoprecipitation experiments (data not shown).
–
It is also intriguing that a search of the literature revealed evidence that activated Snf1 is, in fact, a requirement for Hcm1 nuclear import [22]. It is possible, therefore, that the decreased Snf1 protein abundance in ubc1Δ cells contributes to a negative feed-forward loop, impairing Hcm1 import and ultimately decreasing Fkh1/2 expression and protein abundance. This possibility deserves further investigation, as despite decreased Snf1 abundance, the relative activation state, as measured by its T210 phosphorylation, remains at wild-type levels.
–
We have discovered that the Ub-conjugating enzyme, Ubc1, is indirectly required for SNF1 kinase activity at the level of transcription. Ubc1 function was first associated with the cellular stress response [35], and later with vesicle biogenesis [36], endoplasmic reticulum associated degradation (ERAD) [37], and APC-dependent mechanisms for regulated cell cycle progression [16]. What we describe here appears to be a novel function for Ubc1 that is independent of the APC ubiquitin ligase complex.
–
The schematic cartoon in Fig. 8 highlights our observations and several questions that surround the regulation of SNF1 kinase by Ubc1 activity. Despite WT levels of phosphorylation, nuclear import, and kinase activity on cytosolic and nuclear targets, there remains a deficiency in this activated complex to provide full SUC2 expression. A simple explanation is that this may relate to the relatively low abundance of each of Snf1-Snf4-Gal83 protein subunits found in this heterotrimeric enzyme complex in the Ubc1 deletion strain. This decrease in Snf1 protein levels appears to be indirectly affected by Ubc1 through transcription via the Fkhs and Hcm1 (below) and not through Ub-mediated protein degradation. However, there appears to remain an overriding regulation of Snf1 protein levels that ultimately serves to maintain a stoichiometric balance between the three SNF1 kinase subunits. Secondly, the mechanism underlying the Ubc1-dependent failure of Hmc1 nuclear import is not known. Potential Ubc1 protein targets involved in Hcm1 shuttling are not reported, but may include a cytosolic tether that normally requires Ubc1 for degradation and subsequent release of Hcm1. Alternatively, Ub can serve as a nuclear import signal and this would be a novel role for Ubc1. Next, the enhanced allosterism noted within the SNF1 kinase complex in the ubc1Δ strain may arise from the relative hyper-phosphorylation of Snf1 at Thr210, or from the loss of a Factor X that contributes to steric hindrance of subunit associations. Disruption of the UBA domain in Snf1 also resulted in enhances allosteric associations [11], opening the possibility that this domain or protein face may have protein binding partners that physically regulate SNF1 kinase activity through binding.
MATERIALS AND METHODS
Creation of UBC1 deletion strains
All strains utilized in this manuscript are listed within Table 1. The ubc1∆::KanMX6 cassette was amplified using primers 500 bp up and downstream of UBC1 and genomic DNA isolated from the UBC1/ubc1∆::KanMX6 diploid strain (yTER301) as template. The entire cassette was individually integrated into the Fkh1-TAP, Fkh2-TAP, Mig1-TAP, Rod1-TAP, Snf4-TAP, Gal83-TAP, Hcm1-GFP, Hcm1-TAP, and 2-hybrid reporter strains, with primary selection for successful integrants being KanRes and final confirmation by PCR. Snf1-GFP ubc1Δ was created by crossing (SNF1-GFP::HIS3 x ubc1∆::HIS3), tetrad dissection, scoring for markers, phenotypes, and confirmation by primer-specific PCR amplification.
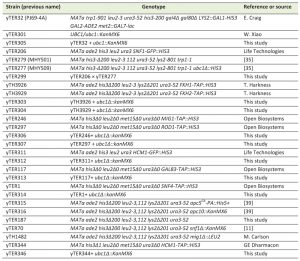 | Table 1. Saccharomyces cerevisiae strains used in this study. |
Total protein extract and western blot analysis
Whole cell protein extracts from logarithmically growing cultures (no growth density greater than OD600 of 0.8) were prepared by a standard bead beat protocol [11] in the presence of RIPA buffer, and protease and phosphatase inhibitors (Sigma). Anti phospho-AMPK (Cell Signaling, 2535L), GFP (Covance, MMS-118P-500), Actin (Sigma, A4700, Lot005134), Clb2 (Santa Cruz, y-180), TAP (Open Biosystems, CAB-1001), and Tubulin (Sigma, 051M4771) antibodies were purchased and the chemiluminescent signal was captured on a VersaDoc (BIO-RAD) molecular imager (Quantity One 4.6.9).
–
Invertase assay
Yeast strains were grown to early log phase in media that was consistent between comparison strains, with all comparison strains consistently in either rich (YPD) or minimal media, according to our published methods [18]. Activity was normalized (value of 1) to that of WT in 0.05% glucose for each biological repeat. Statistical analysis utilized PRISM Version 6.0b software and 2-way ANOVA.
–
Fluorescence microscopy
Fluorescence microscopy was used to detect GFP signal. To determine the subcellular localization of Snf1-GFP and Mig1-GFP, logarithmically growing cultures (OD600 < 0.8) were divided between non-activating complete media (CM), with 2% glucose versus activating (CM 5% glycerol) conditions for 30 minutes. Live cells were moved to mounting medium containing DAPI (Sigma) for immediate DNA visualization. To determine the subcellular localization of Hcm1-GFP, cells were arrested in G1 using α factor addition for 2 hours. Images were taken every 30 minutes following release of G1 arrest. Cells were viewed with an Olympus BX51 fluorescence microscope 100x objective equipped with an Infinity 3-1 UM camera. Images were collected using Infinity Analyse software version 5.0. A minimum of 125 cells for each strain and condition were consecutively scored for co-localization of the GFP-tagged subunits and DAPI nuclear staining.
–
2-hybrid analysis
The yeast WT 2-hybrid reporter strain (PJ69-4α, a gift from S. Fields) and the modified 2-hybrid strain ubc1Δ (ubc1∆::KanMX6 cassette integrated into PJ69-4 α) were doubly transformed with pairs of empty vectors (-ve control: pGAD-C2 and pGBD-C2), or the same backbones expressing unmodified Snf1 and Snf4 subunits (+ve). 1×105 cells from logarithmically growing cultures of each transformation set was repeatedly spotted down the glucose gradient of the slant plates [11], grown at 30°C until colonies were visible, and scanned. Freshly prepared warm liquid X-Gal agarose overlay medium [38] was layered to completely cover cells, solidified and incubated at 30°C and images scanned again after color development.
–
mRNA expression analysis
RNA was isolated (RNAeasy Kit, Qiagen) from logarithmically (OD600 of 0.4) growing WT Snf1-GFP or ubc1Δ Snf1-GFP yeast strains that were transformed with pFkh1-GFP or pFkh2-HA plasmids followed by reverse transcription (QuantiTect Reverse Transcription Kit, Qiagen). cDNA (500 ng) was used as template in amplification reactions and equal reaction volumes were retrieved. Abundance was normalized to signal for ribosomal RNA (rRNA) at 26 cycles. VersaDoc (BIO-RAD) quantitation was obtained from RedSafe nucleic acid stain signal (FroggaBio).
–
Cell cycle arrest
To arrest yeast cells in G1, 4 μg/mL α-factor was added to asynchronous early log phase (OD600 of 0.4) grown in YPD (pH 3.5) prior to experimental start. G1 arrested cell cultures demonstrated >90% of the cells had the expected G1 cell morphology under light microscopy. Cells were released by washing away α-factor and resuspended in fresh YPD. Equal volume cell samples were collected each 30 minutes for protein and fluorescent activated cell sorting (FACS) analysis. . The same Hcm1-GFP strain was used in Fig. 6C and D and Fig. 7A and B.
–
Flow cytometry
Yeast strains of interest were grown in YPD to early logarithmic phase (OD600 of 0.4), 8×105 cells (1 mL) were pelleted and washed with 50 mM Tris buffer (pH 8.0), then resuspended in 1 ml 70% ethanol overnight at room temperature to fix. Cells were pelleted and resuspended in 500 μL of 50 mM Tris buffer and digested with 10 μL 10 mg/mL RNase A at 37°C for 2 hours. Propidium iodide (10 μg/mL) was added to cells for ≥1 hour at room temperature away from light. Flow cytometry was performed using the EPICS® XL and data was analyzed with Flowjo software (v10.0.7).
–
Cycloheximide experiment
To stop protein synthesis, 10 μg/mL cycloheximide was added to live logarithmic phase cells (OD600 < 0.8) in YPD medium. Equal cell numbers were collected every 30 minutes for cell lysate preparation and subsequent western blot analysis.
References
- J. Lu, Y. Lin, J. Sheu, J. Wu, F. Lee, Y. Chen, M. Lin, F. Chiang, T. Tai, S. Berger, Y. Zhao, K. Tsai, H. Zhu, L. Chuang, and J. Boeke, "Acetylation of Yeast AMPK Controls Intrinsic Aging Independently of Caloric Restriction", Cell, vol. 146, pp. 969-979, 2011. http://dx.doi.org/10.1016/j.cell.2011.07.044
- O.E. Owen, G.A. Reichard, M.S. Patel, and G. Boden, "Energy Metabolism in Feasting and Fasting", Advances in Experimental Medicine and Biology, pp. 169-188, 1979. http://dx.doi.org/10.1007/978-1-4757-0734-2_8
- D.G. Hardie, "2. Roles of the AMP-activated/SNFI protein kinase family in the response to cellular stress", Cellular Responses to Stress, pp. 13-28, 1999. http://dx.doi.org/10.1515/9781400865048.13
- J.L. Celenza, and M. Carlson, "A Yeast Gene That Is Essential for Release from Glucose Repression Encodes a Protein Kinase", Science, vol. 233, pp. 1175-1180, 1986. http://dx.doi.org/10.1126/science.3526554
- R. Bergeron, R.R. Russell, L.H. Young, J.M. Ren, M. Marcucci, A. Lee, and G.I. Shulman, "Effect of AMPK activation on muscle glucose metabolism in conscious rats.", The American journal of physiology, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10329989
- C.M. Sutherland, S.A. Hawley, R.R. McCartney, A. Leech, M.J. Stark, M.C. Schmidt, and D. Hardie, "Elm1p Is One of Three Upstream Kinases for the Saccharomyces cerevisiae SNF1 Complex", Current Biology, vol. 13, pp. 1299-1305, 2003. http://dx.doi.org/10.1016/S0960-9822(03)00459-7
- M.A. Treitel, S. Kuchin, and M. Carlson, "Snf1 Protein Kinase Regulates Phosphorylation of the Mig1 Repressor in Saccharomyces cerevisiae", Molecular and Cellular Biology, vol. 18, pp. 6273-6280, 1998. http://dx.doi.org/10.1128/mcb.18.11.6273
- M. Becuwe, N. Vieira, D. Lara, J. Gomes-Rezende, C. Soares-Cunha, M. Casal, R. Haguenauer-Tsapis, O. Vincent, S. Paiva, and S. Léon, "A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis", Journal of Cell Biology, vol. 196, pp. 247-259, 2012. http://dx.doi.org/10.1083/jcb.201109113
- R. Jiang, and M. Carlson, "The Snf1 Protein Kinase and Its Activating Subunit, Snf4, Interact with Distinct Domains of the Sip1/Sip2/Gal83 Component in the Kinase Complex", Molecular and Cellular Biology, vol. 17, pp. 2099-2106, 1997. http://dx.doi.org/10.1128/MCB.17.4.2099
- O. Vincent, R. Townley, S. Kuchin, and M. Carlson, "Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism", Genes & Development, vol. 15, pp. 1104-1114, 2001. http://dx.doi.org/10.1101/gad.879301
- R. Jiao, S. Postnikoff, T.A. Harkness, and T.G. Arnason, "The SNF1 Kinase Ubiquitin-associated Domain Restrains Its Activation, Activity, and the Yeast Life Span", Journal of Biological Chemistry, vol. 290, pp. 15393-15404, 2015. http://dx.doi.org/10.1074/jbc.M115.647032
- K. Hedbacker, "SNF1/AMPK pathways in yeast", Frontiers in Bioscience, vol. 13, pp. 2408, 2008. http://dx.doi.org/10.2741/2854
- M. Zungu, J.C. Schisler, M.F. Essop, C. McCudden, C. Patterson, and M.S. Willis, "Regulation of AMPK by the Ubiquitin Proteasome System", The American Journal of Pathology, vol. 178, pp. 4-11, 2011. http://dx.doi.org/10.1016/j.ajpath.2010.11.030
- M.A. Wilson, E. Koutelou, C. Hirsch, K. Akdemir, A. Schibler, M.C. Barton, and S.Y.R. Dent, "Ubp8 and SAGA Regulate Snf1 AMP Kinase Activity", Molecular and Cellular Biology, vol. 31, pp. 3126-3135, 2011. http://dx.doi.org/10.1128/MCB.01350-10
- K.J. Simpson-Lavy, and M. Johnston, "SUMOylation regulates the SNF1 protein kinase", Proceedings of the National Academy of Sciences, vol. 110, pp. 17432-17437, 2013. http://dx.doi.org/10.1073/pnas.1304839110
- M.C. Rodrigo-Brenni, and D.O. Morgan, "Sequential E2s Drive Polyubiquitin Chain Assembly on APC Targets", Cell, vol. 130, pp. 127-139, 2007. http://dx.doi.org/10.1016/j.cell.2007.05.027
- W. Seufert, J.P. McGrath, and S. Jentsch, "UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation.", The EMBO journal, 1990. http://www.ncbi.nlm.nih.gov/pubmed/2265617
- H. Troy AA, "A Simplified Method for Measuring Secreted Invertase Activity in Saccharomyces cerevisiae", Biochemistry & Pharmacology: Open Access, vol. 03, 2014. http://dx.doi.org/10.4172/2167-0501.1000151
- T. Pramila, W. Wu, S. Miles, W.S. Noble, and L.L. Breeden, "The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle", Genes & Development, vol. 20, pp. 2266-2278, 2006. http://dx.doi.org/10.1101/gad.1450606
- P.C. Hollenhorst, M.E. Bose, M.R. Mielke, U. Müller, and C.A. Fox, "Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae.", Genetics, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10747051
- C. Kühne, and P. Linder, "A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle.", The EMBO journal, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8253070
- M.J. Rodríguez-Colman, M.A. Sorolla, N. Vall-llaura, J. Tamarit, J. Ros, and E. Cabiscol, "The FOX transcription factor Hcm1 regulates oxidative metabolism in response to early nutrient limitation in yeast. Role of Snf1 and Tor1/Sch9 kinases", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1833, pp. 2004-2015, 2013. http://dx.doi.org/10.1016/j.bbamcr.2013.02.015
- B.D. Landry, C.E. Mapa, H.E. Arsenault, K.E. Poti, and J.A. Benanti, "Regulation of a transcription factor network by Cdk1 coordinates late cell cycle gene expression", The EMBO Journal, vol. 33, pp. 1044-1060, 2014. http://dx.doi.org/10.1002/embj.201386877
- D.G. Hardie, D. Carling, and M. Carlson, "THE AMP-ACTIVATED/SNF1 PROTEIN KINASE SUBFAMILY: Metabolic Sensors of the Eukaryotic Cell?", Annual Review of Biochemistry, vol. 67, pp. 821-855, 1998. http://dx.doi.org/10.1146/annurev.biochem.67.1.821
- A. Al-Hakim, A. Zagorska, L. Chapman, M. Deak, M. Peggie, and D. Alessi, "Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains", Biochemical Journal, vol. 411, pp. 249-260, 2008. http://dx.doi.org/10.1042/BJ20080067
- D. Finley, H.D. Ulrich, T. Sommer, and P. Kaiser, "The Ubiquitin–Proteasome System of Saccharomyces cerevisiae", Genetics, vol. 192, pp. 319-360, 2012. http://dx.doi.org/10.1534/genetics.112.140467
- H. Hsu, T. Liu, C. Yeh, T. Chang, Y. Lo, and C. Kao, "Feedback Control of Snf1 Protein and Its Phosphorylation Is Necessary for Adaptation to Environmental Stress", Journal of Biological Chemistry, vol. 290, pp. 16786-16796, 2015. http://dx.doi.org/10.1074/jbc.M115.639443
- K. Elbing, R. McCartney, and M. Schmidt, "Purification and characterization of the three Snf1-activating kinases of Saccharomyces cerevisiae", Biochemical Journal, vol. 393, pp. 797-805, 2006. http://dx.doi.org/10.1042/BJ20051213
- W. Seufert, and S. Jentsch, "In vivo function of the proteasome in the ubiquitin pathway.", The EMBO journal, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1322295
- M.P. Torres, M.J. Lee, F. Ding, C. Purbeck, B. Kuhlman, N.V. Dokholyan, and H.G. Dohlman, "G Protein Mono-ubiquitination by the Rsp5 Ubiquitin Ligase", Journal of Biological Chemistry, vol. 284, pp. 8940-8950, 2009. http://dx.doi.org/10.1074/jbc.M809058200
- F.F. Wiebel, and W. Kunau, "The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes", Nature, vol. 359, pp. 73-76, 1992. http://dx.doi.org/10.1038/359073a0
- M.J. Rodriguez-Colman, G. Reverter-Branchat, M.A. Sorolla, J. Tamarit, J. Ros, and E. Cabiscol, "The Forkhead Transcription Factor Hcm1 Promotes Mitochondrial Biogenesis and Stress Resistance in Yeast", Journal of Biological Chemistry, vol. 285, pp. 37092-37101, 2010. http://dx.doi.org/10.1074/jbc.M110.174763
- M. Muratani, and W.P. Tansey, "How the ubiquitin–proteasome system controls transcription", Nature Reviews Molecular Cell Biology, vol. 4, pp. 192-201, 2003. http://dx.doi.org/10.1038/nrm1049
- D. Hoeller, and I. Dikic, "Targeting the ubiquitin system in cancer therapy", Nature, vol. 458, pp. 438-444, 2009. http://dx.doi.org/10.1038/nature07960
- W. Seufert, and S. Jentsch, "Yeast ubiquitin-conjugating enzymes involved in selective protein degradation are essential for cell viability.", Acta biologica Hungarica, 1991. http://www.ncbi.nlm.nih.gov/pubmed/1844315
- W. Seufert, and S. Jentsch, "Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins.", The EMBO journal, 1990. http://www.ncbi.nlm.nih.gov/pubmed/2154373
- B. Meusser, C. Hirsch, E. Jarosch, and T. Sommer, "ERAD: the long road to destruction", Nature Cell Biology, vol. 7, pp. 766-772, 2005. http://dx.doi.org/10.1038/ncb0805-766
-
A.A. Iniesta, F. García-Heras, J. Abellón-Ruiz, A. Gallego-García, and M. Elías-Arnanz, "Two Systems for Conditional Gene Expression in Myxococcus xanthus Inducible by Isopropyl-β-
d -Thiogalactopyranoside or Vanillate", Journal of Bacteriology, vol. 194, pp. 5875-5885, 2012. http://dx.doi.org/10.1128/JB.01110-12
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
This work was supported by a Natural Sciences and Engineering Research Council (NSERC) Discovery Grant awarded to T.A.
COPYRIGHT
© 2016

The ubiquitin-conjugating enzyme, Ubc1, indirectly regulates SNF1 kinase activity via Forkhead-dependent transcription by Rubin Jiao et al. is licensed under a Creative Commons Attribution 4.0 International License.