Reviews:
Microbial Cell, Vol. 2, No. 4, pp. 94 - 104; doi: 10.15698/mic2015.04.198
Translate to divide: сontrol of the cell cycle by protein synthesis
1 Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, USA.
2 Department of Biology, Texas A&M University, College Station, TX 77843, USA.
Keywords: protein synthesis, ribosome biogenesis, ribosome profiling, START, translational control.
Received originally: 20/01/2015 Received in revised form: 04/03/2015
Accepted: 07/03/2015
Published: 20/03/2015
Correspondence:
Michael Polymenis, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, USA polymenis@tamu.edu
Rodolfo Aramayo, Department of Biology, Texas A&M University, College Station, TX 77843, USA raramayo@tamu.edu
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Michael Polymenis and Rodolfo Aramayo (2015). Translate to divide: Control of the cell cycle by protein synthesis. Microbial Cell 2(4): 94-104.
Abstract
Protein synthesis underpins much of cell growth and, consequently, cell multiplication. Understanding how proliferating cells commit and progress into the cell cycle requires knowing not only which proteins need to be synthesized, but also what determines their rate of synthesis during cell division.
INTRODUCTION
Experiments with proliferating populations of microbial strains, animal or plant cell lines, have rigorous expectations. Under the same culture conditions, cells ought to have the same properties and composition in every single experiment. The basic “metrics” of proliferating cells remain constant, even after many rounds of cell division [1][2]. These metrics include cellular mass and volume, and macromolecular composition [1][3]. The constancy of such parameters reflects the fundamental ability of cells to coordinate their growth with their division [1][4][5]. Balancing cell growth with cell division determines the overall rates of cell proliferation [4][5][6][7][8]. Despite the obvious significance of this phenomenon, how cells manage to coordinate their growth with their division remains largely mysterious.
–
Proteins are often the most abundant macromolecules in proliferating cells. For example, in steady-state cultures of the budding yeast Saccharomyces cerevisiae, the protein content ranges from 35% to 44% of all macromolecules, depending on culture conditions [3]. Furthermore, much of the proteome (>20%) is dedicated to making ribosomes and translation factors, enabling cells to make more proteins [9]. On top of that, making ribosomal components and assembling them into functional ribosomes involves a dizzying array of molecular players and cellular processes [10][11][12]. Consequently, protein synthesis is viewed as a fundamental measure of cell growth. Decades ago, a founding father of cell cycle studies put it this way: “No sensible interpretation of cell growth can be made without a knowledge of the overall pattern of protein synthesis” [13].
–
In the following sections, we discuss the interplay of protein synthesis and cell division. Examples of translational control in embryonic and meiotic cell divisions have been covered comprehensively elsewhere [14][15]. Here, the focus is on mitotic cell division and specifically on the G1 phase of the cell cycle, when cells commit to a new round of cell division. The examples discussed are mainly, but not exclusively, from the budding yeast S. cerevisiae. The discussion centers on un-perturbed, continuously dividing cells, and the impact of genetic, nutritional or chemical perturbations.
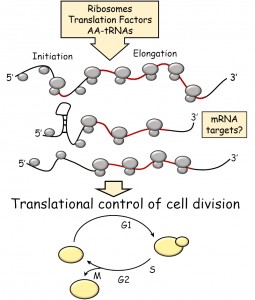 | FIGURE 1: Schematic overview of the topics covered in this review. Open reading frames (ORFs) are shown in red. |
OVERALL PATTERN OF PROTEIN SYNTHESIS IN THE CELL CYCLE
In animal cells, protein synthesis is much lower in mitosis than in other cell cycle phases [16][17][18]. Mechanisms that enable translation of specific mRNAs in animal cells undergoing mitosis have been reviewed elsewhere [15][19]. In contrast to the mitotic block in protein synthesis in animal cells, early studies indicated that budding yeast cells synthesize proteins, including ribosomal proteins, continuously during the cell cycle [20][21][22][23]. These experiments relied mostly on incorporation of labeled amino acids into polypeptides, which were then visualized after electrophoresis [21][22][23]. Hence, those early experiments sampled abundant, constitutively expressed proteins that make up the vast majority of the proteome [9][24][25][26]. Obviously, transcriptional waves drive periodic synthesis of hundreds of proteins in the cell cycle [27][28]. Nonetheless, the bulk of cellular protein synthesis appears to proceed at an exponentially increasing rate in the cell cycle [21][22]. This conclusion was reinforced by monitoring the accumulation of constitutively expressed fluorescent proteins in single cells [29]. In addition, continuous monitoring of cell volume supports an exponential mode of increase in the cell cycle [30]. Therefore, it appears that budding yeast cells make proteins and grow exponentially. Based on buoyant mass as a metric of cell growth, the same can be said about the growth of diverse types of cells, from bacteria to mouse lymphoblasts, with heavier cells growing faster than lighter cells [31]. However, whether or not the growth of animal cells is exponential is still controversial [32][33][34].
–
An exponential mode of protein synthesis and growth is consistent with the existence of active mechanisms that sense some growth metric, perhaps somehow related to protein synthesis [7]. Such mechanisms would enable cells to monitor their growth and commit to a new round of cell division once their growth requirements are met [6][7]. As a result, cells that are born small stay longer in the G1 phase, until they grow enough to commit into and initiate a new round of cell division [4][5][6][7][35]. In yeast, the point of commitment to a new round of cell division is called START [5]. START is marked molecularly by nuclear eviction of the Whi5p repressor [29][36][37], a protein that functions analogously to the retinoblastoma gene product of animal cells [38][39]. Once cells pass through START in late G1, they will initiate and complete their division even if they encounter growth limitations [4][5][35]. To summarize simply, it seems that the bigger yeast cells get, the faster they make proteins and grow, propelling them to divide. This simple concept raises a series of key questions: What determines the rate of protein synthesis? How can the rate of protein synthesis be altered and what would the effects of such alterations be on the cell cycle? What are the RNA targets of translational control that affect cell cycle progression?
INITIATE TO START?
The rate of synthesis of any given protein depends on not only the concentration but also the translational efficiency of its mRNA. Discrepancies between the two parameters underpin translational control. It is often stated that control of translation in eukaryotic cells is exercised mainly at the initiation step, when ribosomes are recruited to mRNA [19]. As discussed in subsequent sections, additional layers of control may also change the rate at which proteins are made. Nonetheless, the initiation step remains a key control point of translation [40][41]. Ribosomal recruitment in eukaryotes usually involves recognition of a cap structure at the 5’-end of the mRNA. The small (40S) ribosomal subunit loaded with initiator Met-tRNA and with the contribution of various initiation factors begins scanning the 5’-UTR of the mRNA for an AUG (or near-cognate start codons). In the process, it has to navigate past the secondary structure of the 5’-UTR [42] or initiation codons upstream of the main open reading frame [43]. Such features may affect recognition and initiation from the correct start codon [40][41][44].
–
The earliest genetic evidence for specific cell cycle effects due to translational control was the isolation of budding yeast conditional mutants in what turned out to be translation initiation factors [5]. One would expect that cessation of a continuous vital cellular function, such as initiation of translation, would simply arrest each cell at whichever point in the cycle that cell happened to be at the time. In an asynchronously proliferating cell population, this would manifest in a pattern of random arrests along the cell cycle [5]. Yet cell division cycle (cdc) genetic screens yielded mutants carrying temperature-sensitive, hypomorphic alleles of translation initiation factors, which did not display a random arrest at their non-permissive temperature. Instead, cells carrying cdc33 (encoding mRNA cap binding protein and translation initiation factor eIF4E [45][46]) or cdc63 (encoding the b subunit of translation initiation factor eIF3 [47][48]) mutations arrest uniformly in the G1 phase of the cell cycle, unable to initiate DNA replication and a new round of cell division [35][46][49][50]. A conditional methionyl-tRNA synthetase (mes1) mutant also arrests in the G1 phase of the cell cycle [51]. These classical genetic analyses suggested strongly that G1 transit is sensitive to translation initiation, more so than other phases of the cell cycle. This conclusion was strengthened when essential gene function was interrogated with a collection of titratable TetO7 promoter alleles for essential genes [52]. In addition to the eIF4E and eIF3b examples mentioned above, Yu et al. showed that inhibiting expression of eIF2a, eIF4A, eIF2b, eIF3i, or eIF1 resulted in G1 arrest in yeast ([52]; and Table 1). Hence, impairing translation initiation in a number of ways, invariably and specifically also impairs the capacity of cells to initiate a new round of cell division.
–
If initiation of translation is important for commitment to division, then signaling pathways that control initiation of division may do so, at least in part, by regulating translation initiation. Mitogenic pathways would be expected to activate translation initiation, while pathways that convey anti-proliferative signals may inhibit translation initiation. The cardinal example for the former case is the Target of Rapamycin (TOR) pathway. How the TOR pathway activates initiation of translation and overall protein synthesis has been reviewed elsewhere [53][54]. Loss of TOR function was known to cause G1 arrest in mammals [55][56] and yeast [53][57]. Connecting the G1 arrest with the effects of TOR on translation, however, was not obvious. In a landmark paper, it was shown that upon loss of TOR function in yeast, the cause of the G1 arrest was a direct consequence of a block in translation initiation [58]. De-repressing translation of the G1 cyclin Cln3p was sufficient to abrogate the G1 arrest of TOR-inhibited yeast cells [58]. TOR is not the only mitogenic pathway that activates translation initiation. The RAS/MAPK pathway in animals phosphorylates and increases the activity of eIF4B [40][59]. Remarkably, phosphorylation of eIF4B on the same residue is a common output of both the TOR and MAPK pathways [59], underscoring the significance of activating translation initiation for commitment to cell division. Conversely, upon stress or starvation it is not prudent to either initiate cell division or make many proteins. It turns out that phosphorylation of eIF2α is a conserved response from yeast to mammals that inhibits overall translation initiation, and it is an output of anti-mitogenic signals [40][41][53][60].
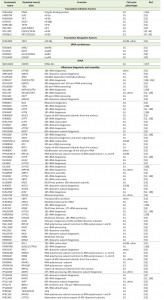 | TABLE 1. Cell cycle phenotypes of loss-of-function mutants in essential genes encoding protein synthesis and ribosome biogenesis factors in S. cerevisiae. |
TRANSLATIONAL TARGETS (?) FOR COMMITMENT TO DIVISION
The above examples suggest that translation initiation goes hand-in-hand with G1 progression and initiation of cell division. By and large, however, they do not answer how this is brought about. What are the relevant proteins important for G1 transit, whose synthesis is sensitive to limitations in translation initiation, and how do these proteins impinge on the machinery of cell division? In the case of the G1 cyclin Cln3p was mentioned above, Hall and colleagues replaced the long 5’-UTR of the yeast CLN3 mRNA with that of UBI4, which is efficiently translated when TOR function is low [58]. Cells carrying this non-repressible CLN3 did not arrest in G1 when TOR function was inhibited by rapamycin [58]. Similarly, efficient translation of CLN3 enabled G1 arrested cdc33 cells, in which the activity of the eIF4E is impaired (see Table 1), to initiate cell division [61]. The Whi3p RNA-binding protein, which sequesters CLN3 mRNA in cytoplasmic foci, may inhibit translation of CLN3 [62]. There is also a uORF in the 5’-UTR of the CLN3 mRNA [63]. We had proposed that the uORF lowers the number of scanning ribosomes that reach the downstream main AUG, especially when the ribosome content of the cell is low in poor media [8][63]. As predicted, inactivation of the uORF in CLN3 allowed cells growing in poor medium, with glycerol as the source for carbon, to accelerate completion of START [63]. Nitrogen limitation was also reported to repress translation of CLN3 [64]. In contrast, another study reported that 20 min after amino acid starvation, translation of CLN3 was up-regulated [43]. While this later discrepancy may simply reflect the different experimental set-ups, there is overall compelling evidence that synthesis of Cln3p, whose levels control the timing of START [65][66], is regulated at least in part at the translational level. It should be noted, however, that all the examples above rest on comparisons between different conditions: with or without inhibitors of TOR [58][63]; mutant vs. wild type [61][62][63]; different nutrients [43][63][64]. It is important to stress that although CLN3 is a translational target, there is no evidence yet that it is targeted in a periodic manner in the cell cycle, in G1 or any other cell cycle phase.
–
It has been reported that CLN3 transcription oscillates early in the cell cycle [67][68], but Cln3p protein levels were not evaluated in these studies. Cln3p is very unstable [69], and difficult to detect by immunoblotting. Early studies reported that Cln3p levels do not oscillate in the cell cycle [70]. Recently, however, more sensitive approaches from two independent studies showed that Cln3p protein is nearly absent in early G1 cells, but it gradually accumulates as cells approach START (Fig. 9 in [71], and Fig. 10 in [72]), without a corresponding increase in the mRNA levels of CLN3. The data from Thorburn et al. [71] and Zapata et al. [72] strongly implicate post-transcriptional mechanisms that control abundance of Cln3p in the cell cycle, perhaps due to control of its synthesis, degradation, or both. Both of these studies relied on centrifugal elutriation to isolate highly synchronous early G1 daughter cells, which were then sampled as they progressed in the cell cycle [71][72]. Until analogous studies are performed with alternative synchronization methods, it is formally possible that the results reflect idiosyncrasies of elutriation. Note also that the CLN3 mRNA cannot possibly be the only physiological target of translational control for cell cycle progression. Cells lacking Cln3p are viable [65][66] and they also respond as expected to nutrient limitations, reducing their size [38].
–
In other systems, the best example of a translational target important for initiation of mitotic cell division is the cyclin-dependent kinase inhibitor p27Kip1 in human cells, whose translation appears to be periodic in the cell cycle, decreasing at the G1/S transition [73][74][75][76][77]. Translational control of p27Kip1 is complex, involving both cap-dependent and independent mechanisms [75][77]. However, because p27Kip1 synthesis decreases at the G1/S transition, this case of translational control cannot account for the postulated activating role of protein synthesis in triggering cell division. Other reported translational targets include the G1 cyclin D1 in mammals [78][79][80], and the G1/S cyclins E1 [81] and E2 [82] in mammals, and Cig2 in fission yeast [83] (reviewed in [84]). Trans-acting factors influencing translation initiation of these targets include the helicase DDX3 for cyclin E1 [81] and Ded1 for Cig2 [83]. It is not clear, however, if translation of these cyclins is periodic in cycling cells. Alternatively, their translational regulation may be an output of a continuous process that affects their overall levels. The levels of these cyclins may oscillate in the cell cycle for other reasons, such as mechanisms that control mRNA levels and protein degradation. Overall, there is a critical gap in our understanding of the role of translational control in mitotic cell cycle progression, especially in G1 progression and commitment to division. There have not been any studies that directly and systematically looked for mRNAs that are translated differentially in the G1 phase, in cycling, un-perturbed cells.
START ON CYCLOHEXIMIDE
After the initiation step, the rate of translation depends on the concentration and activity of translating ribosomes that elongate the nascent polypeptides. If elongation of protein synthesis is inhibited, then what are the consequences on cell cycle progression? This question was first tackled pharmacologically, monitoring cell cycle progression in the presence of varying doses of cycloheximide [35][85][86][87]. Cycloheximide inhibits the translocation step in eukaryotic 80S ribosomes, blocking translational elongation [88]. Increasing doses of cycloheximide increase the population doubling time, mostly because cells spend more time in the G1 phase of the cell cycle [35][85]. Cycloheximide also affects size homeostasis. In budding yeast, cycloheximide reduces the newborn cell size [35][85] and the rate at which cells increase in size [89]. It also increases the critical size threshold for START [85][89]. These changes account for the increase in the duration of the G1 phase upon treatment with cycloheximide. The effects of cycloheximide support the notion that a critical rate of protein synthesis is required for G1 transit and completion of START in budding yeast [86] and animal cells [90][91]. The reports that interrogated cycloheximide’s effects on G1 progression have been influential. They have often been taken to imply a requirement for the continuous synthesis of unstable protein(s), whose rate of synthesis parallels overall protein synthesis rates and cell growth. Identifying those proteins and how they affect the cell division machinery would then hold the promise of explaining molecularly how cells couple their growth with their division [90][91]. Various such candidate proteins have been proposed over the years. However, there is no report of a protein whose levels increase due to cell cycle-dependent translational control, as cycling cells approach START [7].
RIBOSOME MAKES PROTEIN MAKES CELLS?
Ribosomes are the complex macromolecular machines that catalyze protein synthesis. Hence, changing the concentration of functional ribosomes in the cell is expected to change the overall rates of protein synthesis.
–
Bacterial cells grown under conditions that favor fast growth and proliferation (i.e., “rich” media) have more ribosomes than those propagated in “poor” media [92]. These observations suggest that the rate of protein synthesis in bacteria is controlled mostly by ribosome numbers [93]. Although growth rate does not seem to affect significantly the fraction of active ribosomes in the cell (≈80%) or their activity [94], recent observations suggest that even in rich media bacterial ribosomes do not function at maximal elongation rates [95]. Hence, translation rates may be adjusted to proliferation rates by means other than ribosome content.
–
In budding yeast, although protein synthesis rates decrease with decreasing growth rates, there is no proportional decrease in the number of ribosomes per cell [96][97]. For example, a 10-fold drop in growth rate is only accompanied by a 2-fold drop in the number of ribosomes per cell (see Table 4 in [96]). On the other hand, synthesis of ribosomal components is inhibited as cells begin to exhaust the available nutrients and prepare to enter stationary phase [98], or upon amino acid starvation [99]. Apparently, both the activity and the number of ribosomes may be affected in yeast as a function of growth rate.
–
In cycling, unperturbed yeast cells, synthesis of ribosomal components is not cell cycle dependent [10][22][23]. Interfering with ribosome biogenesis, however, affects cell cycle progression dramatically. In yeast, 59 of a total of 78 ribosomal proteins of cytoplasmic ribosomes are encoded by pairs of very similar or identical paralogous genes [100]. Mutants carrying single deletions of those ribosomal protein genes are usually viable [101]. Many of these deletion strains have a small overall cell size [102][103]. A small overall cell size is also characteristic of cells lacking Sfp1p, a transcriptional activator of ribosome biosynthesis and ribosomal protein genes [89]. Cells lacking Sfp1p are born very small [104], and their growth rate is about half of that of wild type cells [71][104]. As a result, the duration of the G1 phase of the cell cycle is greatly increased in sfp1Δ cells [71][89][104]. Still, the smaller critical size threshold of sfp1Δ cells and the smaller overall size of ribosomal protein mutants led Tyers and colleagues to propose that ribosome biogenesis inhibits START in wild type cells [6][89]. In this model, ribosome biogenesis sets the critical size threshold for START, while translation rates of functional ribosomes enable the cells to pass that threshold [6][89]. This model appears paradoxical and counterintuitive, especially since in animals ribosome biosynthesis is thought to promote cell proliferation and cancer [105]. But another finding, that ribosomal proteins may function as haploinsufficient tumor suppressors in animals, appeared to offer support for a negative role of ribosome biogenesis in cell division [106]. However, it was subsequently reported that such effects may be due to cell non-autonomous routes [107]. Furthermore, the following observations argue against the notion that ribosome biogenesis has a general inhibitory role for the initiation of division: First, inhibition of ribosome maturation delays START [108]. Second, despite their small cell size, most ribosomal protein mutants have a longer G1 phase [104][109][110]. Third, loss-of-function of the vast majority of essential ribosome biogenesis factors leads to G1 arrest (see Table 1 and [52]). The most straightforward interpretation of the evidence outlined above is that ribosome biogenesis promotes initiation of cell division in budding yeast.
–
Even when there is a G1 delay due to loss of a ribosomal protein, G1 variables such as birth and critical size of cells lacking individual ribosomal proteins are not uniform, differing qualitatively and quantitatively [104][109][111]. Furthermore, although a delay in G1 is the most common phenotype upon loss of a ribosomal protein, this phenotype is not universal. In several cases, there is no cell cycle phenotype, or a G2/M block is observed instead [104][110]. The basis of all those differences in ribosomal protein mutant phenotypes related to the cell cycle is not clear. Do they reflect specialized translational roles of some ribosomes? Are overall translation rates affected? Is the concentration of ribosomes, their composition, their activity, localization in the cell (or any combination of the above) that is affected? Examples of specialized ribosomal functions abound [112][113][114][115], leading some to speculate on the existence of a “ribosome code” [116]. There are even cases of extraribosomal roles for ribosomal proteins [117]. Regardless of the answers to the above questions, it is imperative that the relevant mRNA substrates affected in each ribosomal protein mutant must be identified, to understand how cell cycle progression might be impacted. However, as is the case for wild type cells (see INITIATE TO START? above), there are no systematic surveys of mRNAs that are translated differentially in the cell cycle in ribosomal protein mutants.
THE OTHER RIBOSOME SUBSTRATES: tRNAS
For the ribosome to elongate at its maximal rate, all its substrates must be present at saturating concentrations. This includes the aminoacyl-tRNAs and the various elongation factors [95]. Inhibition of elongation factor eEF2 may be one way that protein synthesis is inhibited in animal cells in mitosis [118]. However, unlike the numerous examples of perturbations in translation initiation factors that lead to G1 arrest in yeast (Table 1), there is very little analogous evidence for translation elongation factors affecting cell cycle progression. The only reported example is from a large survey of essential genes, reporting that blocking expression of eEF1Bγ in yeast leads to cell cycle arrest in mitosis ([52], see Table 1).
–
Translation elongation rates may be affected by several parameters [119], including the supply and demand for each tRNA. The genetic code is essentially universal. One and the same codon does not code for different amino acids in different organisms. The code, however, is also degenerate. Most amino acids can be specified by more than one codon. A decades-old speculation has been that codons for an amino acid that are used more frequently in an organism would be translated more rapidly than codons for the same amino acid that are rarely used [120]. Differences in codon usage were difficult to test until the advent of ribosome profiling. Sequencing ribosome-protected mRNA fragments allows estimates of ribosome residency on individual codons [43]. Several analyses that included the original ribosome profiling datasets in yeast [43] have examined various parameters, including codon usage, and reached contradictory conclusions [42][119][121][122][123][124][125][126]. Nonetheless, common codons may be translated faster than rare ones in yeast [127]. Modulating which codon is used and the effective concentration of its corresponding tRNA may be a way to influence translation elongation rates both within an mRNA, and across the transcriptome [128]. Surveying codon usage across each codon and tRNA availability revealed that in highly expressed mRNAs rare codons are more prevalent in the first 30-50 codons, slowing down and crowding ribosomes along that stretch [129]. It was proposed that this slow “ramp” is a mechanism that promotes faster translation downstream, by alleviating ribosome “traffic jams” along the mRNA [129]. Furthermore, it turns out that once a particular codon has been used, it will also be used more frequently whenever the same amino acid is encoded downstream in that mRNA [130]. Based on these observations, it was theorized that tRNAs do not diffuse away from the ribosome once they are expelled from it. Instead, the same tRNA for a codon specifying the repeating amino acid is re-charged and channeled for re-use, enabling translation to proceed faster than tRNA diffusion [130].
–
The examples outlined above illustrate how alterations in the supply and demand for each tRNA could impact translation rates. The pertinent question for this review, however, is whether such mechanisms could lead to translational control during the cell cycle. Remarkably, a recent study proposed exactly that kind of regulation, based on codon usage [131]. Optimal codon usage is more prevalent in mRNAs expressed in the G1 phase of the cell cycle, implying that the corresponding mRNAs are translated more efficiently [131]. Interestingly, it has been known for decades that loss-of-function mutations in several tRNA synthetases, and even in a tRNA gene, lead to G1 arrest in yeast (see Table 1). Optimizing translation elongation rates in G1 through codon usage is an exciting possibility. But a correlation is not causation, and the above predictions must await experimental validation, measuring the translational efficiencies of the putative mRNA targets in cycling cells. On a cautionary note, a recent study in mouse embryonic stem cells reported that translation elongation rates were not only independent of codon usage, but also very similar across different mRNAs [132]. Likewise, another study in yeast reported that the rate of translation elongation and translational efficiency were not affected by tRNA abundance, and codon translation rates were not correlated with codon bias [133]. Hence, these issues remain controversial and there might be some time before the dust settles.
DIVIDE TO TRANSLATE?
Implicit in the discussion of all the examples mentioned above is the notion that cell growth and protein synthesis drive cell cycle progression, not the other way around. This is generally the case, established in classic and particularly lucid experiments by Hartwell and colleagues [4][5][35]. However, there are also some changes in the pattern of growth once cell cycle progression is blocked [134][135]. Does that mean that progressing in the cell cycle might also affect the translational control of specific mRNAs? The answer is a resounding yes. Ruggero and colleagues examined by ribosome profiling human cells arrested by thymidine block (G1 and S phases) and nocodazole treatment (G2 phase) in the cell cycle, reporting extensive translational control of numerous mRNAs [17]. Interestingly, this work revealed functional clusters of co-regulated mRNAs. Translational control may be used to coordinate expression of specific cellular machines and processes [17]. These experiments are very important, answering how cell division controls translation. However, they do not answer how translation controls cell division.
OUTLOOK
The role of protein synthesis and translational control has a long history in the cell cycle field. The pioneering experiments of decades ago were incisive, but also largely descriptive. Since then, progress has been incremental and focused on a limited number of putative mRNA translational targets. Identifying all the mRNAs that are under periodic translational control in cycling cells is an obvious and necessary goal. Given the transformative methodologies now available and the current pace of progress, it is only a matter of time before we know how protein synthesis drives cell cycle progression.
References
- F. Neidhardt, J. Ingraham, and M. Schaechter, "Physiology of the bacterial cell.", Sinauer Associates, Inc, Sunderland, Massachusetts., 1990.
- H. Meyer, O. Kappeli, and A. Fiechter, "GROWTH CONTROL IN MICROBIAL CULTURES", Annual Review of Microbiology, vol. 39, pp. 299-319, 1985. http://dx.doi.org/10.1146/annurev.mi.39.100185.001503
- H.C. Lange, and J.J. Heijnen, "Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae", Biotechnology and Bioengineering, vol. 75, pp. 334-344, 2001. http://dx.doi.org/10.1002/bit.10054
- G. JOHNSTON, J. PRINGLE, and L. HARTWELL, "Coordination of growth with cell division in the yeast", Experimental Cell Research, vol. 105, pp. 79-98, 1977. http://dx.doi.org/10.1016/0014-4827(77)90154-9
- J. Pringle, and L. Hartwell, "The Saccharomyces cerevisiae Cell Cycle. The Molecular and Cellular Biology of the Yeast Saccharomyces", Cold Spring Harbor Laboratory Press; pp 97-142., 1981.
- P. Jorgensen, and M. Tyers, "How Cells Coordinate Growth and Division", Current Biology, vol. 14, pp. R1014-R1027, 2004. http://dx.doi.org/10.1016/j.cub.2004.11.027
- J. Turner, J. Ewald, and J. Skotheim, "Cell Size Control in Yeast", Current Biology, vol. 22, pp. R350-R359, 2012. http://dx.doi.org/10.1016/j.cub.2012.02.041
- M. Polymenis, and E.V. Schmidt, "Coordination of cell growth with cell division", Current Opinion in Genetics & Development, vol. 9, pp. 76-80, 1999. http://dx.doi.org/10.1016/S0959-437X(99)80011-2
- W. Liebermeister, E. Noor, A. Flamholz, D. Davidi, J. Bernhardt, and R. Milo, "Visual account of protein investment in cellular functions", Proceedings of the National Academy of Sciences, vol. 111, pp. 8488-8493, 2014. http://dx.doi.org/10.1073/pnas.1314810111
- J.R. Warner, "The economics of ribosome biosynthesis in yeast", Trends in Biochemical Sciences, vol. 24, pp. 437-440, 1999. http://dx.doi.org/10.1016/S0968-0004(99)01460-7
- A. Fatica, and D. Tollervey, "Making ribosomes", Current Opinion in Cell Biology, vol. 14, pp. 313-318, 2002. http://dx.doi.org/10.1016/S0955-0674(02)00336-8
- J. Venema, and D. Tollervey, "Ribosome Synthesis in Saccharomyces cerevisiae", Annual Review of Genetics, vol. 33, pp. 261-311, 1999. http://dx.doi.org/10.1146/annurev.genet.33.1.261
- J. Mitchison, "Cell Growth and Protein Synthesis. The Biology of the Cell Cycle", TCambridge University Press; pp 129., 1971.
- G.A. Brar, M. Yassour, N. Friedman, A. Regev, N.T. Ingolia, and J.S. Weissman, "High-Resolution View of the Yeast Meiotic Program Revealed by Ribosome Profiling", Science, vol. 335, pp. 552-557, 2012. http://dx.doi.org/10.1126/science.1215110
- I. Kronja, and T.L. Orr-Weaver, "Translational regulation of the cell cycle: when, where, how and why?", Philosophical Transactions of the Royal Society B: Biological Sciences, vol. 366, pp. 3638-3652, 2011. http://dx.doi.org/10.1098/rstb.2011.0084
- D. Prescott, and M. Bender, "Synthesis of RNA and protein during mitosis in mammalian tissue culture cells", Experimental Cell Research, vol. 26, pp. 260-268, 1962. http://dx.doi.org/10.1016/0014-4827(62)90176-3
- C. Stumpf, M. Moreno, A. Olshen, B. Taylor, and D. Ruggero, "The Translational Landscape of the Mammalian Cell Cycle", Molecular Cell, vol. 52, pp. 574-582, 2013. http://dx.doi.org/10.1016/j.molcel.2013.09.018
- M.D. Scharff, and E. Robbins, "Polyribosome Disaggregation during Metaphase", Science, vol. 151, pp. 992-995, 1966. http://dx.doi.org/10.1126/science.151.3713.992
- S. Pyronnet, and N. Sonenberg, "Cell-cycle-dependent translational control", Current Opinion in Genetics & Development, vol. 11, pp. 13-18, 2001. http://dx.doi.org/10.1016/S0959-437X(00)00150-7
- D.H. WILLIAMSON, and A.W. SCOPES, "PROTEIN SYNTHESIS AND NITROGEN UPTAKE IN SYNCHRONOUSLY DIVIDING CULTURES OFSACCHAROMYCES CEREVISIAE", Journal of the Institute of Brewing, vol. 67, pp. 39-42, 1961. http://dx.doi.org/10.1002/j.2050-0416.1961.tb01755.x
- S.G. Elliott, and C.S. McLaughlin, "Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae.", Proceedings of the National Academy of Sciences, vol. 75, pp. 4384-4388, 1978. http://dx.doi.org/10.1073/pnas.75.9.4384
- S.G. Elliott, J.R. Warner, and C.S. McLaughlin, "Synthesis of ribosomal proteins during the cell cycle of the yeast Saccharomyces cerevisiae.", Journal of bacteriology, 1979. http://www.ncbi.nlm.nih.gov/pubmed/370092
- R.W. Shulman, L.H. Hartwell, and J.R. Warner, "Synthesis of ribosomal proteins during the yeast cell cycle", Journal of Molecular Biology, vol. 73, pp. 513-525, 1973. http://dx.doi.org/10.1016/0022-2836(73)90097-1
- N. Nagaraj, N. Alexander Kulak, J. Cox, N. Neuhauser, K. Mayr, O. Hoerning, O. Vorm, and M. Mann, "System-wide Perturbation Analysis with Nearly Complete Coverage of the Yeast Proteome by Single-shot Ultra HPLC Runs on a Bench Top Orbitrap", Molecular & Cellular Proteomics, vol. 11, pp. M111.013722, 2012. http://dx.doi.org/10.1074/mcp.M111.013722
- P. Lu, C. Vogel, R. Wang, X. Yao, and E.M. Marcotte, "Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation", Nature Biotechnology, vol. 25, pp. 117-124, 2006. http://dx.doi.org/10.1038/nbt1270
- B. Futcher, G.I. Latter, P. Monardo, C.S. McLaughlin, and J.I. Garrels, "A sampling of the yeast proteome.", Molecular and cellular biology, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10523624
- P.T. Spellman, G. Sherlock, M.Q. Zhang, V.R. Iyer, K. Anders, M.B. Eisen, P.O. Brown, D. Botstein, and B. Futcher, "Comprehensive Identification of Cell Cycle–regulated Genes of the YeastSaccharomyces cerevisiaeby Microarray Hybridization", Molecular Biology of the Cell, vol. 9, pp. 3273-3297, 1998. http://dx.doi.org/10.1091/mbc.9.12.3273
- B. Futcher, "Transcriptional regulatory networks and the yeast cell cycle", Current Opinion in Cell Biology, vol. 14, pp. 676-683, 2002. http://dx.doi.org/10.1016/s0955-0674(02)00391-5
- S.D. Talia, J.M. Skotheim, J.M. Bean, E.D. Siggia, and F.R. Cross, "The effects of molecular noise and size control on variability in the budding yeast cell cycle", Nature, vol. 448, pp. 947-951, 2007. http://dx.doi.org/10.1038/nature06072
- A.K. Bryan, A. Engler, A. Gulati, and S.R. Manalis, "Continuous and Long-Term Volume Measurements with a Commercial Coulter Counter", PLoS ONE, vol. 7, pp. e29866, 2012. http://dx.doi.org/10.1371/journal.pone.0029866
- M. Godin, F.F. Delgado, S. Son, W.H. Grover, A.K. Bryan, A. Tzur, P. Jorgensen, K. Payer, A.D. Grossman, M.W. Kirschner, and S.R. Manalis, "Using buoyant mass to measure the growth of single cells", Nature Methods, vol. 7, pp. 387-390, 2010. http://dx.doi.org/10.1038/nmeth.1452
- I. Conlon, and M. Raff, "Array", Journal of Biology, vol. 2, pp. 7, 2003. http://dx.doi.org/10.1186/1475-4924-2-7
- P. Echave, I.J. Conlon, and A.C. Lloyd, "Cell Size Regulation in Mammalian Cells", Cell Cycle, vol. 6, pp. 218-224, 2007. http://dx.doi.org/10.4161/cc.6.2.3744
- S.S. Grewal, and B.A. Edgar, "Array", Journal of Biology, vol. 2, pp. 5, 2003. http://dx.doi.org/10.1186/1475-4924-2-5
- L.H. Hartwell, and M.W. Unger, "Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division.", The Journal of cell biology, vol. 75, pp. 422-435, 1977. http://dx.doi.org/10.1083/jcb.75.2.422
- U. Eser, M. Falleur-Fettig, A. Johnson, and J. Skotheim, "Commitment to a Cellular Transition Precedes Genome-wide Transcriptional Change", Molecular Cell, vol. 43, pp. 515-527, 2011. http://dx.doi.org/10.1016/j.molcel.2011.06.024
- A. Doncic, M. Falleur-Fettig, and J. Skotheim, "Distinct Interactions Select and Maintain a Specific Cell Fate", Molecular Cell, vol. 43, pp. 528-539, 2011. http://dx.doi.org/10.1016/j.molcel.2011.06.025
- M. Costanzo, J.L. Nishikawa, X. Tang, J.S. Millman, O. Schub, K. Breitkreuz, D. Dewar, I. Rupes, B. Andrews, and M. Tyers, "CDK Activity Antagonizes Whi5, an Inhibitor of G1/S Transcription in Yeast", Cell, vol. 117, pp. 899-913, 2004. http://dx.doi.org/10.1016/j.cell.2004.05.024
- R.A. de Bruin, W. McDonald, T.I. Kalashnikova, J. Yates, and C. Wittenberg, "Cln3 Activates G1-Specific Transcription via Phosphorylation of the SBF Bound Repressor Whi5", Cell, vol. 117, pp. 887-898, 2004. http://dx.doi.org/10.1016/j.cell.2004.05.025
- N. Sonenberg, and A.G. Hinnebusch, "Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets", Cell, vol. 136, pp. 731-745, 2009. http://dx.doi.org/10.1016/j.cell.2009.01.042
- J.W.B. Hershey, N. Sonenberg, and M.B. Mathews, "Principles of Translational Control: An Overview", Cold Spring Harbor Perspectives in Biology, vol. 4, pp. a011528-a011528, 2012. http://dx.doi.org/10.1101/cshperspect.a011528
- M. Kertesz, Y. Wan, E. Mazor, J.L. Rinn, R.C. Nutter, H.Y. Chang, and E. Segal, "Genome-wide measurement of RNA secondary structure in yeast", Nature, vol. 467, pp. 103-107, 2010. http://dx.doi.org/10.1038/nature09322
- N.T. Ingolia, S. Ghaemmaghami, J.R.S. Newman, and J.S. Weissman, "Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling", Science, vol. 324, pp. 218-223, 2009. http://dx.doi.org/10.1126/science.1168978
- H.M. Hood, D.E. Neafsey, J. Galagan, and M.S. Sachs, "Evolutionary Roles of Upstream Open Reading Frames in Mediating Gene Regulation in Fungi", Annual Review of Microbiology, vol. 63, pp. 385-409, 2009. http://dx.doi.org/10.1146/annurev.micro.62.081307.162835
- M. Altmann, and H. Trachsel, "Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cyde division mutant cdc33", Nucleic Acids Research, vol. 17, pp. 5923-5931, 1989. http://dx.doi.org/10.1093/nar/17.15.5923
- C. Brenner, N. Nakayama, M. Goebl, K. Tanaka, A. Toh-e, and K. Matsumoto, "CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae.", Molecular and cellular biology, 1988. http://www.ncbi.nlm.nih.gov/pubmed/3062383
- T. Naranda, S.E. MacMillan, and J.W. Hershey, "Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein.", The Journal of biological chemistry, 1994. http://www.ncbi.nlm.nih.gov/pubmed/7798228
- C. Keierleber, M. Wittekind, S.L. Qin, and C.S. McLaughlin, "Isolation and characterization of PRT1, a gene required for the initiation of protein biosynthesis in Saccharomyces cerevisiae.", Molecular and cellular biology, 1986. http://www.ncbi.nlm.nih.gov/pubmed/3025657
- P.J. Hanic-Joyce, G.C. Johnston, and R.A. Singer, "Regulated arrest of cell proliferation mediated by yeast prt1 mutations", Experimental Cell Research, vol. 172, pp. 134-145, 1987. http://dx.doi.org/10.1016/0014-4827(87)90100-5
- D.P. Bedard, G.C. Johnston, and R.A. Singer, "New mutations in the yeast Saccharomyces cerevisiae affecting completion of ?start?", Current Genetics, vol. 4, pp. 205-214, 1981. http://dx.doi.org/10.1007/BF00420500
- M.W. Unger, and L.H. Hartwell, "Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA.", Proceedings of the National Academy of Sciences, vol. 73, pp. 1664-1668, 1976. http://dx.doi.org/10.1073/pnas.73.5.1664
- L. Yu, L.P. Castillo, S. Mnaimneh, T.R. Hughes, and G.W. Brown, "A Survey of Essential Gene Function in the Yeast Cell Division Cycle", Molecular Biology of the Cell, vol. 17, pp. 4736-4747, 2006. http://dx.doi.org/10.1091/mbc.E06-04-0368
- R. Loewith, and M.N. Hall, "Target of Rapamycin (TOR) in Nutrient Signaling and Growth Control", Genetics, vol. 189, pp. 1177-1201, 2011. http://dx.doi.org/10.1534/genetics.111.133363
- X.M. Ma, and J. Blenis, "Molecular mechanisms of mTOR-mediated translational control", Nature Reviews Molecular Cell Biology, vol. 10, pp. 307-318, 2009. http://dx.doi.org/10.1038/nrm2672
- L.S. Wicker, R.C. Boltz, V. Matt, E.A. Nichols, L.B. Peterson, and N.H. Sigal, "Suppression of B cell activation by cyclosporin A, FK506 and rapamycin", European Journal of Immunology, vol. 20, pp. 2277-2283, 1990. http://dx.doi.org/10.1002/eji.1830201017
- R. Zoncu, A. Efeyan, and D.M. Sabatini, "mTOR: from growth signal integration to cancer, diabetes and ageing", Nature Reviews Molecular Cell Biology, vol. 12, pp. 21-35, 2010. http://dx.doi.org/10.1038/nrm3025
- J. Heitman, N.R. Movva, and M.N. Hall, "Targets for Cell Cycle Arrest by the Immunosuppressant Rapamycin in Yeast", Science, vol. 253, pp. 905-909, 1991. http://dx.doi.org/10.1126/science.1715094
- N.C. Barbet, U. Schneider, S.B. Helliwell, I. Stansfield, M.F. Tuite, and M.N. Hall, "TOR controls translation initiation and early G1 progression in yeast.", Molecular Biology of the Cell, vol. 7, pp. 25-42, 1996. http://dx.doi.org/10.1091/mbc.7.1.25
- D. Shahbazian, P.P. Roux, V. Mieulet, M.S. Cohen, B. Raught, J. Taunton, J.W.B. Hershey, J. Blenis, M. Pende, and N. Sonenberg, "The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity", The EMBO Journal, vol. 25, pp. 2781-2791, 2006. http://dx.doi.org/10.1038/sj.emboj.7601166
- V.A. Cherkasova, and A.G. Hinnebusch, "Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2", Genes & Development, vol. 17, pp. 859-872, 2003. http://dx.doi.org/10.1101/gad.1069003
- P. DANAIE, M. ALTMANN, M.N. HALL, H. TRACHSEL, and S.B. HELLIWELL, "CLN3 expression is sufficient to restore G1-to-S-phase progression in Saccharomyces cerevisiae mutants defective in translation initiation factor eIF4E", Biochemical Journal, vol. 340, pp. 135-141, 1999. http://dx.doi.org/10.1042/bj3400135
- Y. Cai, and B. Futcher, "Effects of the Yeast RNA-Binding Protein Whi3 on the Half-Life and Abundance of CLN3 mRNA and Other Targets", PLoS ONE, vol. 8, pp. e84630, 2013. http://dx.doi.org/10.1371/journal.pone.0084630
- M. Polymenis, and E.V. Schmidt, "Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast", Genes & Development, vol. 11, pp. 2522-2531, 1997. http://dx.doi.org/10.1101/gad.11.19.2522
- C. Gallego, "The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast", The EMBO Journal, vol. 16, pp. 7196-7206, 1997. http://dx.doi.org/10.1093/emboj/16.23.7196
- R. Nash, G. Tokiwa, S. Anand, K. Erickson, and A.B. Futcher, "The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog.", The EMBO journal, 1988. http://www.ncbi.nlm.nih.gov/pubmed/2907481
- F.R. Cross, "DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae.", Molecular and cellular biology, 1988. http://www.ncbi.nlm.nih.gov/pubmed/3062366
- S. Di Talia, H. Wang, J.M. Skotheim, A.P. Rosebrock, B. Futcher, and F.R. Cross, "Daughter-Specific Transcription Factors Regulate Cell Size Control in Budding Yeast", PLoS Biology, vol. 7, pp. e1000221, 2009. http://dx.doi.org/10.1371/journal.pbio.1000221
- V.L. MacKay, B. Mai, L. Waters, and L.L. Breeden, "Early Cell Cycle Box-Mediated Transcription ofCLN3 and SWI4 Contributes to the Proper Timing of the G1-to-S Transition in Budding Yeast", Molecular and Cellular Biology, vol. 21, pp. 4140-4148, 2001. http://dx.doi.org/10.1128/mcb.21.13.4140-4148.2001
- M. Tyers, G. Tokiwa, R. Nash, and B. Futcher, "The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation.", The EMBO journal, 1992. http://www.ncbi.nlm.nih.gov/pubmed/1316273
- M. Tyers, G. Tokiwa, and B. Futcher, "Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins.", The EMBO journal, 1993. http://www.ncbi.nlm.nih.gov/pubmed/8387915
- R.R. Thorburn, C. Gonzalez, G.A. Brar, S. Christen, T.M. Carlile, N.T. Ingolia, U. Sauer, J.S. Weissman, and A. Amon, "Aneuploid yeast strains exhibit defects in cell growth and passage through START", Molecular Biology of the Cell, vol. 24, pp. 1274-1289, 2013. http://dx.doi.org/10.1091/mbc.E12-07-0520
- J. Zapata, N. Dephoure, T. MacDonough, Y. Yu, E.J. Parnell, M. Mooring, S.P. Gygi, D.J. Stillman, and D.R. Kellogg, "PP2ARts1 is a master regulator of pathways that control cell size", Journal of Cell Biology, vol. 204, pp. 359-376, 2014. http://dx.doi.org/10.1083/jcb.201309119
- U. Gopfert, "Cell cycle-dependent translation of p27 involves a responsive element in its 5'-UTR that overlaps with a uORF", Human Molecular Genetics, vol. 12, pp. 1767-1779, 2003. http://dx.doi.org/10.1093/hmg/ddg177
- L. Hengst, and S.I. Reed, "Translational Control of p27 Kip1 Accumulation During the Cell Cycle", Science, vol. 271, pp. 1861-1864, 1996. http://dx.doi.org/10.1126/science.271.5257.1861
- S.S. Millard, A. Vidal, M. Markus, and A. Koff, "A U-Rich Element in the 5′ Untranslated Region Is Necessary for the Translation of p27 mRNA", Molecular and Cellular Biology, vol. 20, pp. 5947-5959, 2000. http://dx.doi.org/10.1128/mcb.20.16.5947-5959.2000
- W.K. Miskimins, G. Wang, M. Hawkinson, and R. Miskimins, "Control of Cyclin-Dependent Kinase Inhibitor p27 Expression by Cap-Independent Translation", Molecular and Cellular Biology, vol. 21, pp. 4960-4967, 2001. http://dx.doi.org/10.1128/mcb.21.15.4960-4967.2001
- M. Kullmann, U. Göpfert, B. Siewe, and L. Hengst, "ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR", Genes & Development, vol. 16, pp. 3087-3099, 2002. http://dx.doi.org/10.1101/gad.248902
- E.J. Campbell Dwyer, H. Lai, R.C. MacDonald, M.S. Salvato, and K.L.B. Borden, "The Lymphocytic Choriomeningitis Virus RING Protein Z Associates with Eukaryotic Initiation Factor 4E and Selectively Represses Translation in a RING-Dependent Manner", Journal of Virology, vol. 74, pp. 3293-3300, 2000. http://dx.doi.org/10.1128/jvi.74.7.3293-3300.2000
- J. Chen, H.E. Feilotter, G.C. Paré, X. Zhang, J.G. Pemberton, C. Garady, D. Lai, X. Yang, and V.A. Tron, "MicroRNA-193b Represses Cell Proliferation and Regulates Cyclin D1 in Melanoma", The American Journal of Pathology, vol. 176, pp. 2520-2529, 2010. http://dx.doi.org/10.2353/ajpath.2010.091061
- Z. Yu, C. Wang, M. Wang, Z. Li, M.C. Casimiro, M. Liu, K. Wu, J. Whittle, X. Ju, T. Hyslop, P. McCue, and R.G. Pestell, "A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation", The Journal of Cell Biology, vol. 182, pp. 509-517, 2008. http://dx.doi.org/10.1083/jcb.200801079
- M. Lai, W. Chang, S. Shieh, and W. Tarn, "DDX3 Regulates Cell Growth through Translational Control of Cyclin E1", Molecular and Cellular Biology, vol. 30, pp. 5444-5453, 2010. http://dx.doi.org/10.1128/mcb.00560-10
- F. Grey, R. Tirabassi, H. Meyers, G. Wu, S. McWeeney, L. Hook, and J.A. Nelson, "A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs", PLoS Pathogens, vol. 6, pp. e1000967, 2010. http://dx.doi.org/10.1371/journal.ppat.1000967
- B. Grallert, S.E. Kearsey, M. Lenhard, C.R. Carlson, P. Nurse, E. Boye, and K. Labib, "A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls.", Journal of cell science, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10725227
- W. Tarn, and M. Lai, "Translational control of cyclins", Cell Division, vol. 6, pp. 5, 2011. http://dx.doi.org/10.1186/1747-1028-6-5
- L. POPOLO, "Control of the yeast cell cycle by protein synthesis*1", Experimental Cell Research, vol. 142, pp. 69-78, 1982. http://dx.doi.org/10.1016/0014-4827(82)90410-4
- S.A. Moore, "Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle.", The Journal of biological chemistry, 1988. http://www.ncbi.nlm.nih.gov/pubmed/3290211
- H.A. Ko, and S.A. Moore, "Kinetic characterization of a prestart cell division control step in yeast. Implications for the mechanism of alpha-factor-induced division arrest.", The Journal of biological chemistry, 1990. http://www.ncbi.nlm.nih.gov/pubmed/2254321
- T.G. Obrig, W.J. Culp, W.L. McKeehan, and B. Hardesty, "The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes.", The Journal of biological chemistry, 1971. http://www.ncbi.nlm.nih.gov/pubmed/5541758
- P. Jorgensen, I. Rupeš, J.R. Sharom, L. Schneper, J.R. Broach, and M. Tyers, "A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size", Genes & Development, vol. 18, pp. 2491-2505, 2004. http://dx.doi.org/10.1101/gad.1228804
- P.W. Rossow, V.G.H. Riddle, and A.B. Pardee, "Synthesis of labile, serum-dependent protein in early G 1 controls animal cell growth", Proceedings of the National Academy of Sciences, vol. 76, pp. 4446-4450, 1979. http://dx.doi.org/10.1073/pnas.76.9.4446
- R. Brooks, "Continuous protein synthesis is required to maintain the probability of entry into S phase", Cell, vol. 12, pp. 311-317, 1977. http://dx.doi.org/10.1016/0092-8674(77)90209-4
- O. Maaløe, and N. Kjeldgaard, "Control of macromolecular synthesis", Control of macromolecular synthesis. W.A. Benjamin, Inc., New York., 1966.
- O. Maaløe, "Regulation of the protein-synthesizing machinery—ribosomes, tRNA, factors, and so on", Biological regulation and development. Springer; pp 487-542., 1979.
- M. Kaczanowska, and M. Rydén-Aulin, "Ribosome Biogenesis and the Translation Process inEscherichia coli", Microbiology and Molecular Biology Reviews, vol. 71, pp. 477-494, 2007. http://dx.doi.org/10.1128/mmbr.00013-07
- M. LOVMAR, and M. EHRENBERG, "Rate, accuracy and cost of ribosomes in bacterial cells", Biochimie, vol. 88, pp. 951-961, 2006. http://dx.doi.org/10.1016/j.biochi.2006.04.019
- C. Waldron, and F. Lacroute, "Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast.", Journal of bacteriology, 1975. http://www.ncbi.nlm.nih.gov/pubmed/1097403
- K.W. Boehlke, and J.D. Friesen, "Cellular content of ribonucleic acid and protein in Saccharomyces cerevisiae as a function of exponential growth rate: calculation of the apparent peptide chain elongation rate.", Journal of bacteriology, 1975. http://www.ncbi.nlm.nih.gov/pubmed/1089627
- Q. Ju, and J.R. Warner, "Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae", Yeast, vol. 10, pp. 151-157, 1994. http://dx.doi.org/10.1002/yea.320100203
- J.R. WARNER, and C. GORENSTEIN, "Yeast has a true stringent response", Nature, vol. 275, pp. 338-339, 1978. http://dx.doi.org/10.1038/275338a0
- I. Wapinski, J. Pfiffner, C. French, A. Socha, D.A. Thompson, and A. Regev, "Gene duplication and the evolution of ribosomal protein gene regulation in yeast", Proceedings of the National Academy of Sciences, vol. 107, pp. 5505-5510, 2010. http://dx.doi.org/10.1073/pnas.0911905107
- G. Giaever, A.M. Chu, L. Ni, C. Connelly, L. Riles, S. Véronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. André, A.P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. Entian, P. Flaherty, F. Foury, D.J. Garfinkel, M. Gerstein, D. Gotte, U. Güldener, J.H. Hegemann, S. Hempel, Z. Herman, D.F. Jaramillo, D.E. Kelly, S.L. Kelly, P. Kötter, D. LaBonte, D.C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S.L. Ooi, J.L. Revuelta, C.J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D.D. Shoemaker, S. Sookhai-Mahadeo, R.K. Storms, J.N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Wang, T.R. Ward, J. Wilhelmy, E.A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J.D. Boeke, M. Snyder, P. Philippsen, R.W. Davis, and M. Johnston, "Functional profiling of the Saccharomyces cerevisiae genome", Nature, vol. 418, pp. 387-391, 2002. http://dx.doi.org/10.1038/nature00935
- P. Jorgensen, J.L. Nishikawa, B. Breitkreutz, and M. Tyers, "Systematic Identification of Pathways That Couple Cell Growth and Division in Yeast", Science, vol. 297, pp. 395-400, 2002. http://dx.doi.org/10.1126/science.1070850
- J. Zhang, C. Schneider, L. Ottmers, R. Rodriguez, A. Day, J. Markwardt, and B.L. Schneider, "Genomic Scale Mutant Hunt Identifies Cell Size Homeostasis Genes in S. cerevisiae", Current Biology, vol. 12, pp. 1992-2001, 2002. http://dx.doi.org/10.1016/s0960-9822(02)01305-2
- S.A. Hoose, J.A. Rawlings, M.M. Kelly, M.C. Leitch, Q.O. Ababneh, J.P. Robles, D. Taylor, E.M. Hoover, B. Hailu, K.A. McEnery, S.S. Downing, D. Kaushal, Y. Chen, A. Rife, K.A. Brahmbhatt, R. Smith, and M. Polymenis, "A Systematic Analysis of Cell Cycle Regulators in Yeast Reveals That Most Factors Act Independently of Cell Size to Control Initiation of Division", PLoS Genetics, vol. 8, pp. e1002590, 2012. http://dx.doi.org/10.1371/journal.pgen.1002590
- D. Ruggero, and P.P. Pandolfi, "Does the ribosome translate cancer?", Nature Reviews Cancer, vol. 3, pp. 179-192, 2003. http://dx.doi.org/10.1038/nrc1015
- A. Amsterdam, K.C. Sadler, K. Lai, S. Farrington, R.T. Bronson, J.A. Lees, and N. Hopkins, "Many Ribosomal Protein Genes Are Cancer Genes in Zebrafish", PLoS Biology, vol. 2, pp. e139, 2004. http://dx.doi.org/10.1371/journal.pbio.0020139
- J.I. Lin, N.C. Mitchell, M. Kalcina, E. Tchoubrieva, M.J. Stewart, S.J. Marygold, C.D. Walker, G. Thomas, S.J. Leevers, R.B. Pearson, L.M. Quinn, and R.D. Hannan, "Drosophila Ribosomal Protein Mutants Control Tissue Growth Non-Autonomously via Effects on the Prothoracic Gland and Ecdysone", PLoS Genetics, vol. 7, pp. e1002408, 2011. http://dx.doi.org/10.1371/journal.pgen.1002408
- K.A. Bernstein, F. Bleichert, J.M. Bean, F.R. Cross, and S.J. Baserga, "Ribosome Biogenesis Is Sensed at the Start Cell Cycle Checkpoint", Molecular Biology of the Cell, vol. 18, pp. 953-964, 2007. http://dx.doi.org/10.1091/mbc.e06-06-0512
- I. Soifer, and N. Barkai, "Systematic identification of cell size regulators in budding yeast", Molecular Systems Biology, vol. 10, 2014. http://dx.doi.org/10.15252/msb.20145345
- M. Thapa, A. Bommakanti, M. Shamsuzzaman, B. Gregory, L. Samsel, J.M. Zengel, and L. Lindahl, "Repressed synthesis of ribosomal proteins generates protein-specific cell cycle and morphological phenotypes", Molecular Biology of the Cell, vol. 24, pp. 3620-3633, 2013. http://dx.doi.org/10.1091/mbc.e13-02-0097
- C. He, S.K. Tsuchiyama, Q.T. Nguyen, E.N. Plyusnina, S.R. Terrill, S. Sahibzada, B. Patel, A.R. Faulkner, M.V. Shaposhnikov, R. Tian, M. Tsuchiya, M. Kaeberlein, A.A. Moskalev, B.K. Kennedy, and M. Polymenis, "Enhanced Longevity by Ibuprofen, Conserved in Multiple Species, Occurs in Yeast through Inhibition of Tryptophan Import", PLoS Genetics, vol. 10, pp. e1004860, 2014. http://dx.doi.org/10.1371/journal.pgen.1004860
- S. Komili, N.G. Farny, F.P. Roth, and P.A. Silver, "Functional Specificity among Ribosomal Proteins Regulates Gene Expression", Cell, vol. 131, pp. 557-571, 2007. http://dx.doi.org/10.1016/j.cell.2007.08.037
- M.N. O'Leary, K.H. Schreiber, Y. Zhang, A.E. Duc, S. Rao, J.S. Hale, E.C. Academia, S.R. Shah, J.F. Morton, C.A. Holstein, D.B. Martin, M. Kaeberlein, W.C. Ladiges, P.J. Fink, V.L. MacKay, D.L. Wiest, and B.K. Kennedy, "The Ribosomal Protein Rpl22 Controls Ribosome Composition by Directly Repressing Expression of Its Own Paralog, Rpl22l1", PLoS Genetics, vol. 9, pp. e1003708, 2013. http://dx.doi.org/10.1371/journal.pgen.1003708
- W.V. Gilbert, "Functional specialization of ribosomes?", Trends in Biochemical Sciences, vol. 36, pp. 127-132, 2011. http://dx.doi.org/10.1016/j.tibs.2010.12.002
- K.K. Steffen, V.L. MacKay, E.O. Kerr, M. Tsuchiya, D. Hu, L.A. Fox, N. Dang, E.D. Johnston, J.A. Oakes, B.N. Tchao, D.N. Pak, S. Fields, B.K. Kennedy, and M. Kaeberlein, "Yeast Life Span Extension by Depletion of 60S Ribosomal Subunits Is Mediated by Gcn4", Cell, vol. 133, pp. 292-302, 2008. http://dx.doi.org/10.1016/j.cell.2008.02.037
- K.B. McIntosh, and J.R. Warner, "Yeast Ribosomes: Variety Is the Spice of Life", Cell, vol. 131, pp. 450-451, 2007. http://dx.doi.org/10.1016/j.cell.2007.10.028
- J.R. Warner, and K.B. McIntosh, "How Common Are Extraribosomal Functions of Ribosomal Proteins?", Molecular Cell, vol. 34, pp. 3-11, 2009. http://dx.doi.org/10.1016/j.molcel.2009.03.006
- S. White-Gilbertson, D.T. Kurtz, and C. Voelkel-Johnson, "The role of protein synthesis in cell cycling and cancer", Molecular Oncology, vol. 3, pp. 402-408, 2009. http://dx.doi.org/10.1016/j.molonc.2009.05.003
- T. Tuller, I. Veksler-Lublinsky, N. Gazit, M. Kupiec, E. Ruppin, and M. Ziv-Ukelson, "Composite effects of gene determinants on the translation speed and density of ribosomes", Genome Biology, vol. 12, pp. R110, 2011. http://dx.doi.org/10.1186/gb-2011-12-11-r110
- J.B. Plotkin, and G. Kudla, "Synonymous but not the same: the causes and consequences of codon bias", Nature Reviews Genetics, vol. 12, pp. 32-42, 2010. http://dx.doi.org/10.1038/nrg2899
- M. Siwiak, and P. Zielenkiewicz, "A Comprehensive, Quantitative, and Genome-Wide Model of Translation", PLoS Computational Biology, vol. 6, pp. e1000865, 2010. http://dx.doi.org/10.1371/journal.pcbi.1000865
- T. Tuller, Y.Y. Waldman, M. Kupiec, and E. Ruppin, "Translation efficiency is determined by both codon bias and folding energy", Proceedings of the National Academy of Sciences, vol. 107, pp. 3645-3650, 2010. http://dx.doi.org/10.1073/pnas.0909910107
- W. Qian, J. Yang, N.M. Pearson, C. Maclean, and J. Zhang, "Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency", PLoS Genetics, vol. 8, pp. e1002603, 2012. http://dx.doi.org/10.1371/journal.pgen.1002603
- C.A. Charneski, and L.D. Hurst, "Positively Charged Residues Are the Major Determinants of Ribosomal Velocity", PLoS Biology, vol. 11, pp. e1001508, 2013. http://dx.doi.org/10.1371/journal.pbio.1001508
- E.W. Wallace, E.M. Airoldi, and D.A. Drummond, "Estimating Selection on Synonymous Codon Usage from Noisy Experimental Data", Molecular Biology and Evolution, vol. 30, pp. 1438-1453, 2013. http://dx.doi.org/10.1093/molbev/mst051
- J. Yang, X. Chen, and J. Zhang, "Codon-by-Codon Modulation of Translational Speed and Accuracy Via mRNA Folding", PLoS Biology, vol. 12, pp. e1001910, 2014. http://dx.doi.org/10.1371/journal.pbio.1001910
- J. Gardin, R. Yeasmin, A. Yurovsky, Y. Cai, S. Skiena, and B. Futcher, "Measurement of average decoding rates of the 61 sense codons in vivo", eLife, vol. 3, 2014. http://dx.doi.org/10.7554/elife.03735
- K. Fredrick, and M. Ibba, "How the Sequence of a Gene Can Tune Its Translation", Cell, vol. 141, pp. 227-229, 2010. http://dx.doi.org/10.1016/j.cell.2010.03.033
- T. Tuller, A. Carmi, K. Vestsigian, S. Navon, Y. Dorfan, J. Zaborske, T. Pan, O. Dahan, I. Furman, and Y. Pilpel, "An Evolutionarily Conserved Mechanism for Controlling the Efficiency of Protein Translation", Cell, vol. 141, pp. 344-354, 2010. http://dx.doi.org/10.1016/j.cell.2010.03.031
- G. Cannarozzi, N.N. Schraudolph, M. Faty, P. von Rohr, M.T. Friberg, A.C. Roth, P. Gonnet, G. Gonnet, and Y. Barral, "A Role for Codon Order in Translation Dynamics", Cell, vol. 141, pp. 355-367, 2010. http://dx.doi.org/10.1016/j.cell.2010.02.036
- M. Frenkel‐Morgenstern, T. Danon, T. Christian, T. Igarashi, L. Cohen, Y. Hou, and L.J. Jensen, "Genes adopt non‐optimal codon usage to generate cell cycle‐dependent oscillations in protein levels", Molecular Systems Biology, vol. 8, 2012. http://dx.doi.org/10.1038/msb.2012.3
- N. Ingolia, L. Lareau, and J. Weissman, "Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes", Cell, vol. 147, pp. 789-802, 2011. http://dx.doi.org/10.1016/j.cell.2011.10.002
-
C. Pop, S. Rouskin, N.T. Ingolia, L. Han, E.M. Phizicky, J.S. Weissman, and D. Koller, "Causal signals between codon bias,
mRNA structure, and the efficiency of translation and elongation", Molecular Systems Biology, vol. 10, 2014. http://dx.doi.org/10.15252/msb.20145524 - A.I. Goranov, and A. Amon, "Growth and division—not a one-way road", Current Opinion in Cell Biology, vol. 22, pp. 795-800, 2010. http://dx.doi.org/10.1016/j.ceb.2010.06.004
- A.I. Goranov, M. Cook, M. Ricicova, G. Ben-Ari, C. Gonzalez, C. Hansen, M. Tyers, and A. Amon, "The rate of cell growth is governed by cell cycle stage", Genes & Development, vol. 23, pp. 1408-1422, 2009. http://dx.doi.org/10.1101/gad.1777309
ACKNOWLEDGMENTS
Work in the authors’ laboratories has been supported with grants from the National Science Foundation (MCB-0818248 to M.P.) and the National Institutes of Health (GM58770 to R.A.). The open access publishing fees for this article have been covered by the Texas A&M University Online Access to Knowledge (OAK) Fund, supported by the University Libraries and the Office of the Vice President for Research.
COPYRIGHT
© 2015

Translate to divide: сontrol of the cell cycle by protein synthesis by Michael Polymenis and Rodolfo Aramayo is licensed under a Creative Commons Attribution 4.0 International License.









