Research Articles:
Microbial Cell, Vol. 6, No. 11, pp. 509 - 523; doi: 10.15698/mic2019.11.697
Transcriptomic and chemogenomic analyses unveil the essential role of Com2-regulon in response and tolerance of Saccharomyces cerevisiae to stress induced by sulfur dioxide
1 Universidade de Trás-os-Montes e Alto Douro, Escola de Ciências da Vida e Ambiente; Vila Real, Portugal.
2 BioISI – Biosystems & Integrative Sciences Institute, Campo Grande, Lisboa, Portugal.
3 Life and Health Sciences Research Institute (ICVS), School of Medicine, University of Minho, 4710-057 Braga, Portugal.
4 ICVS/3B’s – PT Government Associate Laboratory, Braga/Guimarães, Portugal.
5 Institute for Bioengineering and Biosciences, Instituto Superior Técnico, Department of Bioengineering, University of Lisbon, Portugal.
Keywords: Sulfur dioxide tolerance, Com2 (YER130c), wine preservation, Saccharomyces cerevisiae, stress response.
Abbreviations: ROS – reactive oxygen species.
Received originally: 09/07/2019 Received in revised form: 04/09/2019
Accepted: 13/09/2019
Published: 30/09/2019
Correspondence:
Ana Mendes-Ferreira, , BioISI - Universidade de Trás-os-Montes e Alto Douro, Escola de Ciências da Vida e Ambiente; Vila Real, Portugal; anamf@utad.pt
Nuno P Mira, Instituto Superior Técnico, Department of Bioengineering, University of Lisbon, Portugal; nuno.mira@tecnico.ulisboa.pt
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Patrícia Lage, Belém Sampaio-Marques, Paula Ludovico, Nuno P Mira and Ana Mendes-Ferreira (2019). Transcriptomic and chemogenomic analyses unveil the essential role of Com2-regulon in response and tolerance of Saccharomyces cere-visiae to stress induced by sulfur dioxide. Microbial Cell 6(11): 509-523. doi: 10.15698/mic2019.11.697
Abstract
During vinification Saccharomyces cerevisiae cells are frequently exposed to high concentrations of sulfur dioxide (SO2) that is used to avoid overgrowth of unwanted bacteria or fungi present in the must. Up to now the characterization of the molecular mechanisms by which S. cerevisiae responds and tolerates SO2 was focused on the role of the sulfite efflux pump Ssu1 and investigation on the involvement of other players has been scarce, especially at a genome-wide level. In this work, we uncovered the essential role of the poorly characterized transcription factor Com2 in tolerance and response of S. cerevisiae to stress induced by SO2 at the enologically relevant pH of 3.5. Transcriptomic analysis revealed that Com2 controls, directly or indirectly, the expression of more than 80% of the genes activated by SO2, a percentage much higher than the one that could be attributed to any other stress-responsive transcription factor. Large-scale phenotyping of the yeast haploid mutant collection led to the identification of 50 Com2-targets contributing to the protection against SO2 including all the genes that compose the sulfate reduction pathway (MET3, MET14, MET16, MET5, MET10) and the majority of the genes required for biosynthesis of lysine (LYS2, LYS21, LYS20, LYS14, LYS4, LYS5, LYS1 and LYS9) or arginine (ARG5,6, ARG4, ARG2, ARG3, ARG7, ARG8, ORT1 and CPA1). Other uncovered determinants of resistance to SO2 (not under the control of Com2) included genes required for function and assembly of the vacuolar proton pump and enzymes of the antioxidant defense, consistent with the observed cytosolic and mitochondrial accumulation of reactive oxygen species in SO2-stressed yeast cells.
INTRODUCTION
Sulfur dioxide (often abbreviated to sulfite or SO2) has been long used in winemaking due to its recognized potential in inhibiting growth of spoilage microbes whose activity decreases wine quality by producing off-flavor compounds (e.g. H2S and other sulfur-based volatiles) and causing formation of sediments or gas after bottling, among other deleterious effects. SO2 is usually added to the must in the form of potassium or sodium metabisulfite that, in solution, has a pH dependent speciation. At low pH the more abundant species is molecular SO2 (pKa ~ 1.8), however, at the pH of wine (between 3 and 3.8) bisulfite (HSO3−; pKa 6.9) is the more abundant form [1]. The antimicrobial potential of SO2 is believed to result from its ability to permeate the microbial plasma membrane by passive diffusion [2], similar to what is known to occur with carboxylic weak organic acids also used as preservatives (e.g. acetic or propionic acids) [3]. Once inside microbial cells molecular SO2 dissociates into bisulfite (HSO3−) and sulfite (SO32-) due to the internal pH surpassing the pKa values [1]. The deleterious effects caused by accumulation of SO2, HSO3− or SO32- inside Saccharomyces cerevisiae cells were described to include depletion of ATP caused by inhibition of glyceraldeyde-3-phosphate dehydrogenase and alcohol dehydrogenases [4], perturbation of the plasma membrane structure and damaging of proteins, vitamins or coenzymes due to cleavage of disulfide bonds [5][6]. Although significant strain-to-strain variability is observed concerning tolerance to SO2, studies have identified spoilage yeast strains able to grow in the presence of concentrations as high as 600 mg/L [7][8][9], well above the levels legally permitted in enology which are in the range of 150 to 400 mg/L [10]. Among the more feared SO2-tolerant species are the yeasts Saccharomycodes ludwigii and Brettanomyces bruxellensis, with the first being particularly problematic due to the difficulties in eradicating it from contaminated environments using currently available sanitation methodologies [11]. As a response to that emergence in resilience to SO2 within spoilage species, wine producers tend to increase the concentration of this preservative. This practice has, however, adverse effects in health of more susceptible consumers [12][13], while also rendering the wines less attractive in a market that demands “chemical-free” products and increasing pressure for selection of more tolerant strains.
–
Wine S. cerevisiae strains are considerably more tolerant to SO2 than laboratory strains and genomic analysis revealed that, in most cases, this phenotype results from a higher transcription of the SSU1 gene [14][15][16][17][18][19], encoding a plasma membrane pump essential for efflux of sulfite and bisulfite [20]. This increased transcription of SSU1 observed in SO2-tolerant wine strains results from chromosomal rearrangements that exchange the native promoter for a stronger one [14][15][16][17][18]. The more common of these rearrangements is a translocation between chromosome XVI (where the SSU1 allele is natively located) and chromosome VIII, placing SSU1 under the regulation of the strong promoter of ECM34 [14][15][17]. Notably, SSU1 transcription is not responsive to SO2 [16][18] and thus the different levels of tolerance in S. cerevisiae wine strains are, in general, explained by the basal transcript levels of SSU1 [16][18]. An exception to this was observed for the 71B commercial strain, which was found to encode a SO2-responsive SSU1 allele, presumably due to the retention of the original SSU1 promoter in addition to two copies of SSU1-R [21]. The transcription factor Fzf1 is the regulator of SSU1 expression from its endogenous promoter [20], however, it plays no role in the control of the higher expression of more tolerant strains since it has no binding site in the modified SSU1 promoters these strains harbor [15][18]. Besides the efflux of sulfite mediated by Ssu1, the production of acetaldehyde, the increased activity of the transulfuration pathway and the blockage of adenine biosynthesis have also been pinpointed as mechanisms contributing to the improved tolerance of S. cerevisiae to SO2 [22][23][24].
–
In this work, we demonstrate that the poorly characterized transcription factor Com2 (ORF YER130c) is essential for tolerance and response of S. cerevisiae cells to SO2 at pH 3.5. Com2 encodes an orphan homologue of the environmental stress-responsive transcription factors Msn2 and Msn4 [25], being derived from a separate locus in the ancestral yeast genome that existed before whole genome duplication [26]. Com2 was also found to be an orthologue of Candida albicans Mnl1, a species also harboring an orthologue of S. cerevisiae Msn4 but not of Msn2 [26]. CaMsn4 and CaMnl1 could not be implicated in a general environmental stress response in C. albicans [26] but CaMnl1 was found to mediate tolerance and response to high concentrations of acetic acid [26]. Although transcription of Com2 was found to increase in S. cerevisiae cells exposed to acetic, propionic or benzoic acids (at pH 4) [27][28][29], no protective effect against these acids could be attributed to this regulator [27][28][29]. By exploring a combination of transcriptomics and genome-wide phenotypic analyses, a detailed mechanism describing how Com2-dependent regulon contributes for tolerance and response of S. cerevisiae to SO2 is established in this study. The results also provided, for the first time, a genome-wide view of the S. cerevisiae genes required for maximal tolerance to SO2 at a low pH broadening the current view into the modes of toxicity of this preservative and underlying protective responses.
–
RESULTS
The transcription factor Com2 is a determinant of S. cerevisiae tolerance to SO2
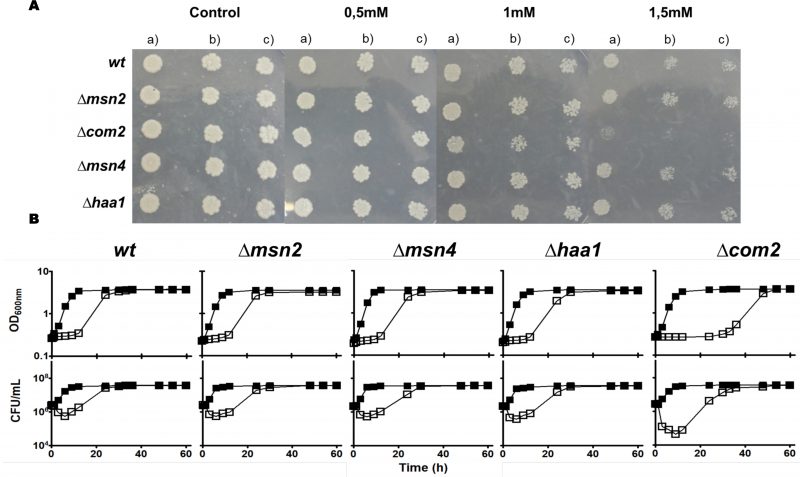
The demonstrated similarity of Com2 with Msn2/Msn4 [26] and the reported involvement of the latter two regulators in S. cerevisiae response to environmental stress, specifically to stress induced by carboxylic acids [30], prompted us to compare the susceptibility of mutants devoid of COM2, MSN2 or MSN4 to SO2 (at pH 3.5) with the one of the parental strain BY4741 (Fig. 1). Since Haa1 was also implicated in tolerance to hydrophilic organic acids [27] and is a positive regulator of Com2 [28][29], a mutant devoid of this gene was also included in this phenotypic profiling (Fig. 1). Among the strains examined only com2Δ showed strong susceptibility to SO2 (at pH 3.5), this being visible both in solid and in liquid medium (Fig. 1A). Consistently, cellular viability of SO2-challenged com2Δ cells was well below the one registered for any of the other strains, this difference being particularly noticeable during the adaptation period (Fig. 1B). These results suggest that Com2 plays an essential role in the early adaptation phase of S. cerevisiae to stress imposed by SO2 at pH of 3.5. We tested whether deletion of Com2 would provide tolerance to other stresses including toxic concentrations of ethanol, copper or of H2O2 with no significant protective effect being observed (results not shown).
–
–
General overview on the yeast transcriptional response to SO2 in BY4741 and in BY4741_com2Δ cells
The identification of Com2 as a new determinant of yeast tolerance to SO2 led us to investigate its role in the transcriptome-wide alterations imposed by stressful concentrations of that preservative. For this, we have resorted to a transcriptomic profiling of S. cerevisiae BY4741 and com2Δ cells cultivated for 60 min in the absence or presence of 0.5 mM SO2 (at pH 3.5), these being experimental conditions comparable to those used in the susceptibility assays shown in Fig. 1. The selected time-point for the transcriptomic analysis corresponds to the early phase of adaptation to SO2 (Figure S1), a period where strong adaptive responses to carboxylic acid-induced stresses have been observed [28][29] and where Com2 seems to exert a more prominent protective effect (Fig. 1B).
–
Principal component analysis of the results obtained for the different samples used in the transcriptomic analysis revealed that exposure to SO2 greatly impacted genomic expression in both wild-type and com2Δ yeast cells, although this effect was markedly different for the two strains (Figure S1). Comparison of the transcriptomes of wild-type and com2Δ cells grown in the absence of SO2 (control conditions) revealed only 48 genes with significantly different expression in the two strains, 28 of these being more expressed (above 2-fold) in the wild-type strain and 20 being more expressed in the mutant strain (using the same 2-fold threshold). The list of these genes affected upon COM2 deletion in control conditions is provided in Table S1.
–
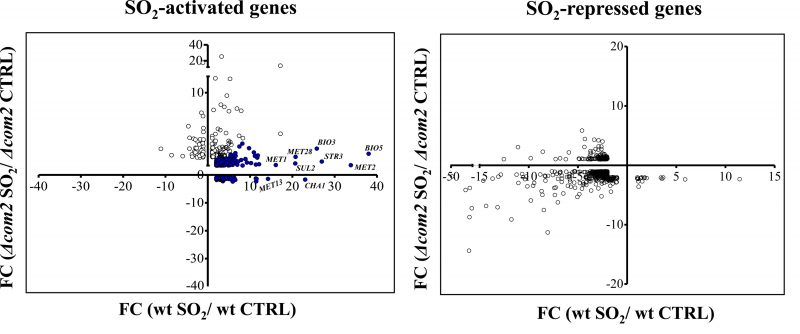
Exposure to SO2 resulted in a dramatic change in the genomic expression of BY4741 cells with 569 genes being up-regulated and 456 genes down-regulated, comparing with the levels attained in control conditions (listed in Table S2). In the com2Δ strain, exposure to SO2 resulted in the activation of 161 genes and in repression of 242, however, these genes had little overlap with those that changed their expression in the wild-type strain (see data and figure in Table S3). The differences in the genomic expression programs prompted by both strains in response to SO2 are rendered clear in Fig. 2. A high number of genes activated by SO2 in wild-type cells were not over-expressed in the mutant background and, in some cases, these genes were even repressed (Fig. 2). These SO2-responsive genes lacking activation in the mutant are highlighted in blue in Fig. 2 and were considered as Com2-targets. In concerns to genes down-regulated by SO2, a higher similarity was observed for wild-type and com2Δ cells, although it was still possible to observe genes that were down-regulated in the wild-type but not in the mutant and vice versa (Fig. 2). However, because Com2 is predicted as a transcriptional activator, we did not consider the genes differently repressed by SO2 in the wild-type and in the com2Δ mutant as being Com2-targets.
–
–
To render the toxic effects of SO2 at a low pH in the physiology of S. cerevisiae BY4741 cells clearer, the genes found to be up- and down-regulated were clustered according with their predicted biological function using the MIPS functional catalogue. The biological functions enriched in the dataset of SO2-induced genes are “amino acid metabolism”, “nitrogen and sulfur metabolism”, “metabolism of vitamins and prosthetic groups”, “ion transport”, “amino acid transport” and “interaction with the environment”, as detailed in Figure S2. In general, these functional classes coincide with those obtained in the prior transcriptomics analyses of S. cerevisiae SO2-stressed cells [31][32] and also of C. albicans [33]. Despite this, our number of SO2-responsive genes was much higher than the one reported in other studies performed in S. cerevisiae [31][32], which can be attributed to the higher sensitivity of the BY4741 strain in comparison with the susceptibility of the wine strains explored in these other studies [31][32].
–
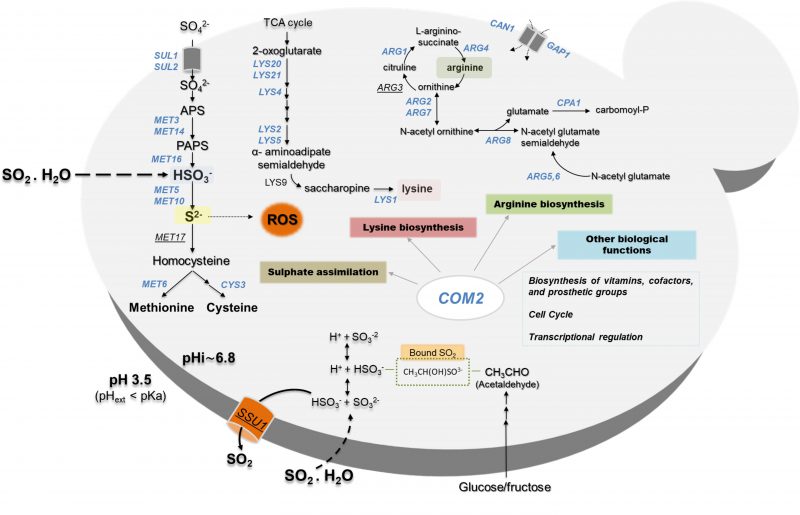
A closer look into the data revealed that the “amino acid metabolism” and “nitrogen and sulfur metabolism” classes were largely composed of genes involved in the sulfate assimilation pathway (SUL1, SUL2, MET3, MET14, MET16, MET5, MET10) as well as genes involved in biosynthesis of lysine (LYS20, LYS21, LYS4, LYS2, LYS9 and LYS1) and arginine (ARG2, ARG3, ARG7, ARG4, ARG5/6, CPA1 and ARG81) (Table S1). Consistent with their observed up-regulation, deletion of MET3, MET5, MET10, MET14, MET16, LYS4, LYS14, LYS20, LYS21, ARG2, ARG3, ARG4, ARG5, ARG7 genes led to a strong susceptibility phenotype to SO2 at pH 3.5 (Fig. 3). Previously, the addition of methionine to the medium was found to increase susceptibility of yeast cells to SO2 by inhibiting the activity of the sulfate assimilation pathway [22]. Under the conditions that we have tested this does not seem to be the case since susceptibility to SO2 of the BY4741 strain (auxotrophic for methionine) was identical to the one of the prototrophic yfgΔ0 strain which was cultivated in a growth medium not supplemented with methionine (compare results in Figures.S1, S3 and Fig. 1). Furthermore, the susceptibility phenotype of the met5Δ, met14Δ and met16Δ mutants to SO2 was still clearly detectable in the prototrophic background (Figure S3). Supplementation of the MMB medium with lysine (1.0 g/L) alleviated SO2 toxicity (Fig. 3), while in the case of arginine a positive effect was also observed but only for the highly susceptible strain com2Δ (results not shown).
–
–
Consistent with the reported lack of transcriptional regulation of SSU1 by SO2 in S. cerevisiae [15][31][32], we could not detect the up-regulation of this sulfite exporter neither in the transcriptomic profiling performed nor in other additional experiments that were performed using different cell extracts (Figure S4). Genes involved in acetaldehyde synthesis were also not found to be activated in response to SO2, despite the description that increased synthesis of this sulfur-sequestering carbohydrate ameliorate the toxic effects of SO2 [23][24]. Nonetheless, reduced expression of genes encoding enzymes involved in degradation of acetaldehyde into ethanol or acetate was detected (Table 1), which could indicate an attempt to increase (or at least maintain) the internal concentration of acetaldehyde [24].
–
TABLE 1. List of genes whose SO2-induced transcriptional activation registered in S. cerevisiae BY4741 cells is abolished in the absence of COM2 |
| TABLE 1: List of genes whose SO2-induced transcriptional activation registered in S. cerevisiae BY4741 cells is abolished in the absence of COM2. |
Genes whose elimination leads to an increase in yeast susceptibility to SO2 are highlighted in gray. |
–
Defining the Com2-regulon in response to SO2
Deletion of COM2 abolished the transcriptional activation of 503 genes (these genes being highlighted in blue in Fig. 2) and reduced, by more than 50%, the SO2-induced activation of other 15 genes. It is important to highlight that although a short-exposure time to SO2 had been used to identify the Com2-regulated genes, more specifically responding to that preservative, it is likely that this high number of genes might not result because they are specific targets of Com2 in response to SO2 but rather because they reflect a different physiological state of the much more susceptible mutant cells, in comparison with the wild-type cells. A subset of these SO2-induced genes up-regulated by Com2 is shown in Table 1 and the full list is provided in Table S4. From the functional point of view, this set of Com2-targets was highly enriched for genes involved in sulfate assimilation (MET8, MET10, MET3, MET5, MET14, MET1, MET22 and MET16) and transport (SUL1 and SUL2), as well as other genes associated with methionine and cysteine metabolism and regulation (MET32, SAM2, HOM3, MET13 MET1, MHT1, SAM1, MET4, STR2, MET30 and STR3). Also included is this regulon are genes involved in biosynthesis of lysine (LYS2, LYS21, LYS20, LYS14, LYS4, LYS5, LYS1 and LYS9), arginine (ARG5,6, ARG4, ARG2, ARG3, ARG7, ARG8, ORT1 and CPA1) or biotin (BIO2, VHR1, ISA1, BIO5, BIO4 and BIO3), a sulfur containing vitamin. The positive effect of Com2 in up-regulating some of these SO2-responsive genes was confirmed by RT-PCR (Figure S4). Twelve of these genes up-regulated by Com2 under SO2 stress (PHO5, SAM1, Ho, SAM2, USB1, OPT1, MET13, YHP1, MET6, PSD1, MOG1 and WSC4) already showed a lower expression in the mutant under control conditions, although the impact of COM2 deletion in the expression of these genes is significantly higher in the presence of SO2 (Table S1).
–
Genome-wide profiling of SO2-resistance genes: role of genes of the Com2-regulon
The strong susceptibility phenotype exhibited by com2Δ cells to SO2 may result from the reduced expression of genes required for maximal tolerance to that compound. Indeed, the protective effect against SO2 of the Com2-regulated genes MET3, MET5, MET10, MET14, MET16, ARG2, ARG3, ARG4, ARG5/6, ARG7, ARG8, LYS4, LYS14, LYS20 and LYS21 was demonstrated in this work (Fig. 3A and B). However, the very large size of the Com2-regulon renders difficult to individualize the individual contribution of the remaining target genes in tolerance to SO2 and therefore we have resorted to the utilization of the yeast deletion mutant collection for a broader phenomic analysis. In the phenotypic profiling undertaken all the 5,000 haploid yeast strains that are devoid of all non-essential genes (generally known as the yeast disruptome) were profiled for their growth in MMB solid medium supplemented with 1, 1.5 or 2 mM of SO2 (at pH 3.5). This screening revealed around 767 mutants with reduced growth in the presence of SO2, comparing with the one exhibited by wild-type cells, 282 being hyper-susceptible (unable to grow at 1 mM) and 485 susceptible (did not show growth at 1.5 mM or 2 mM) (Table S5). Around 100 of the identified SO2-susceptible strains are devoid of genes providing protection against multi-stresses in yeast (highlighted in grey in Table S5) [34] and therefore their identification in this screening was expected. Comparison of the identified SO2-resistance genes with those that were found to be transcriptionally activated in response to this chemical revealed a small overlap of only 50 genes (highlighted in grey in Table S5). This observation is in line with previous reports describing a poor correlation between yeast genes induced in response to environmental stressors (including to stress induced by carboxylic acids) and those providing protection against these stresses [3][28][29][35]. Crossing the set of defined Com2 targets with the list of genes providing tolerance to SO2 resulted in the identification of 47 genes (highlighted in grey in Table S5 and in Table 1). Besides the Com2-targets involved in biosynthesis of lysine and in the sulfate assimilation pathway that were mentioned above, the other Com2-targets contributing to SO2 tolerance uncovered by the phenomic analysis included genes involved in transcriptional regulation (e.g. ZAP1, BAS1, URE2), in regulation of cell cycle (e.g. BUD14, BUD16, DMA2 or KIP2) or in transport (e.g. MCH5, STP22 or MDM34).
–
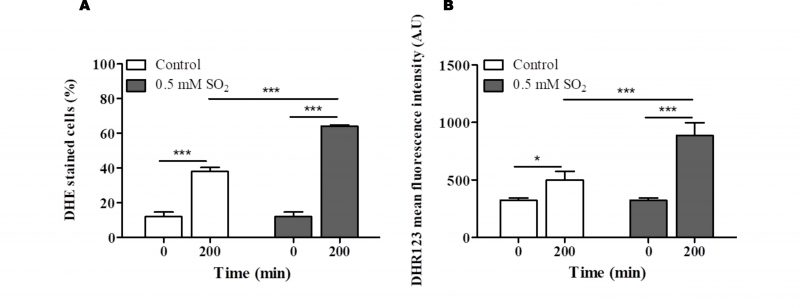
Functional clustering of the determinants of SO2 resistance uncovered in this work (shown in Fig. S5) reinforced the relevance of genes involved in biosynthesis of arginine, methionine and lysine (see Fig. 3), as well of genes required for biosynthesis of adenine (e.g. BAS1, ADE2), glycine, serine or histidine (e.g. GVC1, SER33, SER1, SER2) for maximal tolerance to this preservative (Table S5). Genes involved in synthesis of the reserve carbohydrates trehalose and glycogen (e.g. GPH1, TSL1, TPS1, TPS2, TPS3) as well as the key glycolytic enzymes Pfk1, Tdh3 and Tpi1 were also identified as being critical determinants of SO2 tolerance (Table S5 and Fig. S5), presumably by contributing to generate enough energy to allow the cells to activate the necessary defense mechanisms. Consistent with this idea, inactivation of glycolysis has been pinpointed as one of the relevant mechanisms of toxicity of sulfite in yeast [4] and genes required for assembly of components of the electron transfer (COQ5, COX11, COX14, COX15 or COX23) and of F1F0-ATPase function (e.g ATP1, ATP11, ATP12 or ATP15) were also found to provide protection against SO2 (supplementary Table S5). Another detectable feature was the identification of all genes required for assembly and function of the V-ATPase (VMA1, VMA11, VMA16, VMA21, VMA3, VMA4, VMA5, VMA7 or VPS1) as being essential for tolerance to SO2 (Table S5 and Figure. S5). SO2 was found to induce intracellular acidification [3] and therefore the requirement for V-ATPase is likely to be related with the demonstrated involvement of this pump in restoring internal pH to physiological values upon acid-induced stress [2][24]. Several genes involved in antioxidant defense were also found to provide protection against SO2 including the cytosolic superoxide dismutase SOD1, the peroxiredoxin AHP1, the reductases GRX3 or MXR2 and the glutathione biosynthetic enzyme GSH1 (Table S5). These observations suggest that one of the features of SO2 exposure is the induction of oxidative stress, a hypothesis that was confirmed in SO2-challenged cells by the higher accumulation of reactive oxygen species (ROS) in the cytosol, as revealed by the higher fluorescence intensity levels of DHR123, and of the ROS superoxide anion, revealed by the higher number of cells stained with DHE (Fig. 4). In line with these observations, a mutant devoid of cytosolic superoxide dismutase has been found to have increased susceptibility to sulfite (also in line with the results from our disruptome screening), this being hypothesized to result from an oxyradical-based mechanism of toxicity of this chemical [36]. Sulfite has been found to induce oxidative stress in kidney, cardiac and plant cells [37][38], the mechanism behind this being hypothesized to result from the perturbation of mitochondrial function mainly through the inhibition of cytochrome c activity by sulfide accumulated upon sulfite reduction [37]. Several genes encoding components of the cytochrome c oxidase complex were found to have a reduced expression in response to SO2 in S. cerevisiae, with CYC7 exhibiting a particularly prominent repression reaching almost 50-fold (Table S2).
–
–
DISCUSSION
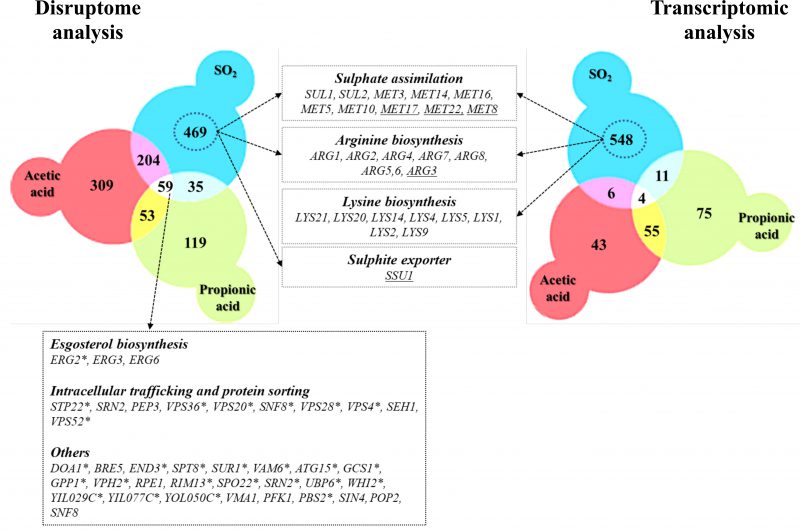
In this work, the responses of S. cerevisiae cells to SO2 at the enological relevant pH of 3.5 were investigated integrating results from transcriptomic and chemogenomic analyses. Although the mode of action of SO2 was associated with the one of weak carboxylic organic acids [2][24], comparison of the genes up-regulated by SO2 with those responding to acetic or propionic acids (chosen for this comparative analysis for being more hydrophilic, like sulfite) reveals a modest overlap (Fig. 5). The set of genes found to provide protection against acetic or propionic acids also showed little similarity with those required for maximal tolerance to SO2 (Fig. 5) and SO2 induced the Com2-dependent regulon while acetic and propionic acids rely on the Haa1-dependent regulons [27][28][29]. Altogether, these observations suggest that once inside yeast cells, SO2, acetic and propionic acids exert dissimilar toxic effects and, consequently, the responses evolved by yeast cells are different and require different players. The fact that SO2 is not organic is likely to be on the basis of these anticipated toxic effects in the cellular environment. The genes of the sulfate activating pathway emerged among the SO2-specific transcriptional responses as well as among the SO2-specific determinants of resistance (SO2) (Fig. 5). Prior studies have referred to the relevance of the sulfate activation pathway in yeast response to SO2 [22][32], however, this was mostly based on the observed higher expression of these genes in more tolerant strains [22]. Interestingly, up-regulation of genes of the sulfate activation pathway in response to sulfite stress was observed in C. albicans and in Vitis vinifera [37][38] indicating that responsiveness of this pathway to sulfite/SO2 stress is not unique to S. cerevisiae. In the present study, we not only confirmed that the transcription of genes of the sulfate activating pathway is highly responsive to SO2 stress, but we also showed that these genes are critical for tolerance. The protective effect of the sulfite reductases Met5 and Met10 goes in line with the anticipated involvement of these enzymes in reducing the intracellularly accumulated sulfite into sulfide [22][32], which can then be channeled to synthesis of methionine and cysteine (as schematically represented in Fig. 6). However, the requirement for Met3, Met14 or Met16 tolerance to SO2 is much less obvious since these enzymes are located upstream of the sulfite-sulfide reduction step (Fig. 6). In plants the involvement of the sulfate reduction steps in response to sulfite was linked with an increase in intracellular sulfate caused by sulfite oxidation via a sulfite oxidase [37]. We cannot exclude oxidation of intracellularly accumulated sulfite into sulfate or another species (e.g. sulfonic acid), which could require detoxification through the sulfate activating pathway, however, we do not favor this hypothesis since sulfite oxidases were never described in S. cerevisiae (although it has been shown in Rhodotorula [39]). Sulfite was found to degrade thiamine [40] and thus one may hypothesize that it may also have a deleterious effect against other sulfur-containing molecules like cysteine, methionine or homocysteine. Under these conditions, SO2-stressed cells would require a high flux through the sulfate activating pathway as a compensatory response, something that could not be achieved in the met3Δ, met14Δ or met16Δ mutants (thus explaining their susceptibility phenotype). In mammalian cells a similar hypothesis has been raised based on the idea that synthesis of cysteine and cystine (the oxidized form of cysteine) can serve as a scavenging mechanism for excess sulfite [41]. Further studies are required to test this hypothesis and mechanistically understand what is the precise role of the sulfate activating pathway in SO2 detoxification by yeast cells, although it seems clearer that it goes beyond the conversion of exceeding sulfite into sulfide.
–
–
The involvement of Com2 in response and tolerance of S. cerevisiae cells to stress induced by SO2 at low pH is the first biological function attributed to this regulator. Transcriptomic analysis revealed that Com2 regulates, directly or indirectly, approximately 80% of the SO2-activated genes. According with the information available in the YEASTRACT database [42], this percentage of targets is much higher than the one predicted for other stress-responsive transcription factors (e.g. Msn2/Msn4 had only 47% of documented targets among the SO2-activated genes; Figure S6 and data not shown). These results clearly highlight Com2 as the main player in the reprogramming of yeast genomic expression under SO2 stress. In total, 47 genes activated by Com2 in response to SO2 were found to contribute for maximal tolerance to this chemical including not only the genes of the sulfate activating pathway but also genes required for lysine and arginine biosynthesis, among others that have more general functions in response to environmental stress. Consistently, wine fermentations undertaken in the presence of SO2 rapidly exhaust the arginine available in the must (comparing with the consumption observed in the absence of SO2) [43][44], this also being observed for lysine although at a much less significant extent [43].
–
–
Most of the SO2-activated genes that are documented targets of Msn2 were also found to be regulated by Com2 suggesting some overlap between these two regulatory systems (Figure S6). The DNA sequence recognized by Com2 is not yet known, however, the C-terminal region of this regulator is highly similar to the DNA binding domains of ScMsn2 and CaMnl1 (Figure S7) which prompted us to search the promoter region of Com2-targets for STRE-like motifs (5'-CCCCT-3'=), known to serve as binding sites for ScMsn2 and CaMnl1 [26]. A high percentage of the SO2-induced genes identified as targets of Com2 and Msn2 harbor in their promoter region STRE-like sequences (Figure S6), however, these were not abundant on the promoters of the genes specifically under the regulation of Com2 (Figure S6). This leads us to conclude that either Com2 recognizes a STRE-like sequence and the effect observed on the regulation of genes only regulated by this transcription factor is indirect and mediated via other regulators; or Com2 recognize a sequence less specific than STRE.
–
In the absence of SO2 Com2 had no significant effect on the yeast transcriptome suggesting that it becomes active only when cells are exposed to this chemical. In line with this, up to now only IME1 had been identified as a Com2 target [45] regardless the extensive number of studies describing transcriptional reprogramming of yeast cells under various stress conditions. Recently, several yeast stress-responsive transcription factors have been found to become activated upon binding of xenobiotics this being reported, for example, for the drug-induced regulator Pdr1 [46] as well as for Haa1 that was found to directly bind acetate [47]. Further studies are required to demonstrate if Com2 becomes activated in response to SO2 as the result of binding sulfite or metabisulfite. A shift in cytoplasmic to nuclear localization may also be another hypothesis as a regulatory mechanism for Com2 activity, considering that this was described to regulate ScMsn2 activity [48][49][50][51], however, the localization signals for nuclear import (NLS) and export (NES) mapped in Msn2 (between 575-642 residues) [52] are not conserved in Com2 (Fig. S7) and no other NLS or NES can be bioinformatically predicted for this regulator (Figure S7). Although Com2 had been identified as the orphan-homologue of C. albicans Mnl1 they do not appear to respond to the same stimuli since Com2 is dispensable for tolerance and response to acetic acid [27][29], while Mnl1 is essential for this response [26]. Furthermore, no significant over-representation of genes of the Mnl1-regulon was observed in a recent survey of C. albicans response to sulfite stress [33].
–
A model of how S. cerevisiae cells respond at a genome-wide level to SO2-induced stress is proposed in Fig. 6? integrating the data that we have gathered in our work with previously established knowledge. The central role of Com2 in SO2 response and tolerance is highlighted in this scheme (Fig. 6?), showing its involvement in the up-regulation of newly uncovered determinants of resistance like the genes of the sulfate activating pathway or the genes required for biosynthesis of lysine and arginine. It is possible that SO2-induced stress can lead to a depletion of intracellular lysine and arginine which can justify the protective effect exerted by enzymes contributing to the synthesis of these two amino acids as well, however, further studies are required to clarify this. Interestingly, recently it was shown that an increase in intracellular arginine improves yeast tolerance to inhibitory concentrations of ethanol by contributing to the integrity of the cell wall and the plasma membrane [53], which are also two well-described targets of SO2-induced toxicity according to the results of our chemogenomics screening. Notably, while supplementation of the MMB medium with lysine led to improved growth in the presence of SO2, no significant effect was obtained upon arginine supplementation which can result from a difficulty in increasing the intracellular concentration of this amino acid just by increasing its availability in the growth medium since the arginine uptake is complex and involves many players whose regulation of activity can be affected by the experimental conditions, including the exposure to SO2. Nonetheless, the demonstration that lysine supplementation can alleviate the toxic effects of SO2 in S. cerevisiae represents an important finding of our work since it can be easily explored by winemakers to improve performance of yeast strains along vinification, especially those that are more susceptible to the preservative action of SO2.
–
It is expected that the knowledge gathered in this work can be used for the development of more efficient wine preservation strategies based on SO2, either helping to understand what can be the molecular mechanisms behind the highly tolerant spoilage yeasts B. bruxellensis or S. ludwigii or by guiding the identification of what are the adaptive responses necessary to increase robustness of S. cerevisiae strains to be used in vinification. The herein identified set of genes involved in response and resistance to SO2 may also pave the way for a better understanding of what can be the toxicity mechanisms exerted by this preservative for humans in a more toxicological perspective.
–
MATERIALS AND METHODS
Strains and growth media
The parental strain Saccharomyces cerevisiae BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and the 19 individual single deletion mutant strains (yer130cΔ, msn2Δ, msn4Δ, haa1Δ, met3Δ, met14Δ, met16Δ, met5Δ, met10Δ, arg2Δ, arg3Δ, arg4Δ, arg5,6Δ, arg7Δ, arg8Δ, lys4Δ, lys21Δ, lys14Δ and lys20Δ) used in this work were all acquired from Euroscarf. The prototrophic strain S. cerevisiae yfgΔ0 (MATα::KanMX can1Δ::STE2prΔSpHIS5 his3Δ1 lyp1Δ0) as well as the derived mutants met14Δ, met16Δ and met5Δ were kindly provided by Dr. Amy Caudy [54]. The different yeast strains were cultivated in YPD medium (containing, per liter, 20g glucose (Merck), 10 g bactopeptone (HiMedia) and 5 g yeast extract (HiMedia)) or in minimal? medium MMB (containing, per liter, 1.7 g YNB without amino acids or ammonium sulfate (Difco Laboratories, Detroit, Michigan), 20 g glucose (Merck) and 2.65 g (NH4)2SO4 (Merck)). To surpass the auxotrophies of the BY4741 background, the MMB medium was further supplemented with 20 mg/L methionine, 60 mg/L leucine, 20 mg/L histidine, 20 mg/L uracil, all acquired from Sigma (Spain). Whenever required the pH of the MMB medium was adjusted to 3.5 using HCl as the acidulant. Preparation of the corresponding solid YPD or MMB media was achieved upon supplementation with 2% agar (Merck).
–
Sulfur dioxide susceptibility assays
Susceptibility to SO2 of the parental strain BY4741 or of the selected deletion mutants was compared using spot assays or through the comparison of growth curves in liquid medium. For the spot assays, mid-exponential phase cells (OD600nm ~ 0.6) cultivated in liquid MMB medium (at pH 3.5) at 30°C with orbital agitation (250 rpm), were used to prepare suspensions (in distilled water) having an OD600nm of 0.1. These suspensions and two subsequent dilutions (1:5 and 1:10) were applied as spots (4 µl) onto the surface of solid MMB (pH 3.5) supplemented with SO2 concentrations ranging from 0 to 1.5 mM. SO2 was included in media by incorporating equal amounts of freshly prepared stock solutions of potassium metabisulfite (Merck) in water, with pH adjusted to 3.5, to give the desired final concentrations. Plates were incubated at 30°C for 2 to 3 days, depending on the severity of growth inhibition. For the comparison of the growth curves in liquid medium, mid-exponential phase cells (OD600nm ~ 0.6) cultivated in MMB medium (at pH 3.5) were used to inoculate fresh medium at an initial OD600nm of 0.2. Growth was followed by accompanying the increase in OD600nm of the culture as well as the concentration of viable cells (assessed as the number of colony forming units per ml of cell culture (CFU ml-1) onto YPD solid medium, after 2 days of incubation at 30°C. Alternatively, growth of the wild-type and of the selected mutants in the presence or absence of SO2 was accompanied in 96-multiwell plates for 24h. The experimental setting used in this case was identical to the one described above with the exception that cultures were incubated in a plate reader (Multiskan Ascent spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA)) and OD600nm readings were taken every 30 min. All experiments were carried out in, at least, triplicates. To test the effect of lysine and arginine supplementation (1 g/L) cells were cultured in 96-multiwell plates as described above.
–
Transcriptomic analysis of S. cerevisiae BY4741 and BY4741_com2Δ in response to SO2 stress
S. cerevisiae BY4741 and the mutant devoid of COM2 were cultivated in MMB growth medium (at pH 3.5) until mid-exponential phase (standard OD600nm 0.6) and then re-inoculated in fresh medium, either or not supplemented with 0.5 mM of SO2 (at pH 3.5). After 60 min of incubation in the absence or presence of SO2, cells were harvested, immediately frozen in liquid nitrogen, and kept at – 80°C until total RNA extraction. Total RNA extraction was performed according to the hot phenol method. Concentration and purity were determined by spectrophotometry and integrity was confirmed using an Agilent 2100 Bioanalyzer with a RNA 6000 Nano Assay (Agilent Technologies, Palo Alto, CA, USA). RNA was processed for use on Affymetrix (Santa Clara, CA, USA) GeneChip Yeast Genome 2.0 Arrays, according to the manufacturer's One-Cycle Target Labeling Assay. Briefly, 5 mg of total RNA containing spiked in Poly-A RNA controls (GeneChip Expression GeneChip Eukaryotic Poly-A RNA Control Kit; Affymetrix) was used in a reverse transcription reaction (One-Cycle DNA synthesis kit; Affymetrix) to generate first-strand cDNA. After second strand synthesis, double-stranded cDNA was used in an in vitro transcription (IVT) reaction to generate biotinylated cRNA (GeneChip Expression 30-Amplification Reagents for IVT-Labeling; Affymetrix). Size distribution of the cRNA and fragmented cRNA, respectively, was assessed using an Agilent 2100 Bioanalyzer with a RNA 6000 Nano Assay. A total of 5 μg of fragmented cRNA was used in a 100-μL hybridization cocktail containing added hybridization controls. 80 μl of mixture was hybridized on arrays for 16 h at 45 ºC. Standard post hybridization wash and double-stain protocols (FS450_0003; GeneChip HWS kit) were used on an Affymetrix GeneChip Fluidics Station 450. Arrays were scanned on an Affymetrix Gene-Chip scanner 3000 7G.
–
The scanned arrays were analyzed first with Affymetrix MAS 5.0 software to obtain Absent/Present calls and subsequently with DNA-Chip Analyzer (dChip) 2010 (http://www.dchip.org, Wong Lab, Harvard). The arrays were normalized to a baseline array with median CEL intensity by applying an Invariant Set Normalization Method [40]. Normalized CEL intensities of the eight arrays were used to obtain model-based gene expression indices based on a PM (Perfect Match)-only model [40]. Microarray data are available from Gene Expression Omnibus (GEO) with accession number GSE 117883. Genes were considered to be differentially expressed if they were called Present in at least two replicates of each sample. A lower-confidence bound fold change cut-off between experiments and baseline was above 1.2, maintain a false discovery rate (FDR) below 10%. For downstream analysis of SO2-responsive genes only the genes whose transcriptional change was above or below 2-fold (p-value below 0.05) the values registered in control cells were considered to be altered. Clustering of the up- or down-regulated genes, based on biological function, was performed using the MIPS Functional Catalogue and the description of gene function is based on the information available in Saccharomyces Genome Database. To confirm some of the results obtained in the microarray analysis carried out, the transcript levels of selected SO2-responsive genes were compared by real-time RT-PCR in BY4741 and com2Δ cells. The same experimental setting used for the microarray analysis was used. Synthesis of cDNA from total RNA samples was performed using the MultiscribeTM reverse transcriptase kit and the subsequent real time RT-PCR step was carried out using SYBR® Green reagents 7500 RT-PCR in a Thermal Cycler Block (Applied Biosystems, Bedford, MA, USA). Specific primers for the amplification of the selected genes and of the ACT1 gene, used as an internal control, were designed using Primer Express Software (Applied Biosystems) (available upon request).
–
Phenotypic screening of the yeast deletion mutant collection to SO2-induced stress
To screen the Euroscarf haploid yeast mutant collection the strains were grown for 16h in 96-wells plate in MMB medium (at pH 3.5) at 30°C with orbital agitation (250 rpm). Five replicates of the wild type strain BY4741 were included in each plate to minimize inter and intra experimental condition variation. Then, 3 µL of the cellular suspensions were spotted (using a 96-pin replica platter) onto the surface of MMB solid medium (at pH 3.5) supplemented with 0, 1 or 1.5 mM SO2. Yeast cells growth was visually inspected after incubation at 30ºC during 3 days. Strains showing a growth defect in control plates, without SO2, were discarded. Strains showing reduced growth comparing to the one obtained for the wild-type strain in the presence of 1 mM SO2 were considered highly susceptible, while those showing a growth defect only in the presence of 1.5 were considered susceptible.
–
Assessment of intracellular reactive oxygen species in SO2-challenged cells
Cells of the parental strain BY4741 were cultivated, at 30ºC with orbital agitation (250 rpm), in MMB (pH 3.5) until exponential-phase and then re-inoculated into this same fresh medium supplemented with 0 (control) or 0.5 mM of SO2 (at pH 3.5). After 200 minutes of incubation at 30 ºC (maintaining an orbital agitation of 250 rpm) cells were stained with dihydroethidium (DHE, Molecular Probes, Eugene, USA) or with dihydrorhodamine 123 (DHR123, Molecular Probes, Eugene, USA) to determine intracellular reactive oxygen species (ROS), as described in (Mesquita A et al., 2010). DHE (10 µg ml-1) was added to the cell suspensions and these were further incubated for 10 min in dark. For the DHR123 staining, 15 µg ml-1 of DHR123 were added and cells incubated for 90 minutes at 30°C in the dark. Fluorescence emitted by DHE or DHR123 was analyzed by LSRII flow cytometer (BD-Biosciences) with a 488 nm excitation laser. The DHE and DHR123 signals were collected through a 488-nm blocking filter, with a 685-nm long-pass or a 505-nm long-pass, respectively. 30,000 cells/sample were captured at a low flow rate of 1,000 cells/s. Data collected with the LSRII flow cytometer were processed with Flowjo software (Tree Star).
–
REFERENCES
- Corte L, Roscini L, Zadra C, Antonielli L, Tancini B, Magini A, Emiliani C, Cardinali G (2012). Effect of pH on potassium metabisulphite biocidic activity against yeast and human cell cultures. Food Chem 134(3): 1327-1336. doi: 10.1016/j.foodchem.2012.03.025
- STRATFORD M, MORGAN P, ROSE AH (1987). Sulphur Dioxide Resistance in Saccharomyces cerevisiae and Saccharomycodes ludwigii. Microbiology 133(8): 2173-2179. doi: 10.1099/00221287-133-8-2173
- Mira NP, Teixeira MC, Sa-Correia I (2010). Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 14(5): 525-540. doi: 10.1089/omi.2010.0072
- Hinze H, Holzer H (1986). Analysis of the energy metabolism after incubation of Saccharomyces cerevisiae with sulfite or nitrite. Arch Microbiol 145(1): 27-31. doi: 10.1007/bf00413023
- Romano P, Suzzi G (1993). Sulphur dioxide and wine microorganisms. In: Fleet G, editor Wine Microbiology and Biotechnology.Harwood, Amsterdam; pp 373–393.
- Fugelsang K, Edwards C (2007). Wine Microbiology. Springer, US. doi: 10.1007/978-0-387-33349-6
- Conterno L, Joseph C, Arvik T, Henick-Kling T, Bisson L (2006). Genetic and Physiological Characterization of Brettanomyces bruxellensis Strains Isolated from Wines. Am J Enol Vitic 57: 139–147.
- Curtin C, Kennedy E, Henschke PA (2012). Genotype-dependent sulphite tolerance of Australian Dekkera (Brettanomyces) bruxellensis wine isolates. Lett Appl Microbiol 55(1): 56-61. doi: 10.1111/j.1472-765X.2012.03257.x
- Vigentini I, Joseph CM, Picozzi C, Foschino R, Bisson LF (2013). Assessment of the Brettanomyces bruxellensis metabolome during sulphur dioxide exposure. FEMS Yeast Res 13(7): 597-608. doi: 10.1111/1567-1364.12060
- Wine IOoVa (2018). International Code of Oenological Practice. Available at http://www.oiv.int/en/technical-standards-and-documents/oenological-practices/international-code-of-oenological-practices [Accessed 07.07.2019]
- Vejarano R (2018). Saccharomycodes ludwigii, Control and Potential Uses in Winemaking Processes. Fermentation 4(3): 71. doi: 10.3390/fermentation4030071
- Irwin SV, Fisher P, Graham E, Malek A, Robidoux A (2017). Sulfites inhibit the growth of four species of beneficial gut bacteria at concentrations regarded as safe for food. PLoS One 12(10): e0186629. doi: 10.1371/journal.pone.0186629
- Vally H, Misso NL (2012). Adverse reactions to the sulphite additives. Gastroenterol Hepatol Bed Bench 5(1): 16-23. PMID: 24834193
- Perez-Ortin JE, Querol A, Puig S, Barrio E (2002). Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res 12(10): 1533-1539. doi: 10.1101/gr.436602
- Yuasa N, Nakagawa Y, Hayakawa M, Iimura Y (2005). Two alleles of the sulfite resistance genes are differentially regulated in Saccharomyces cerevisiae. Biosci Biotechnol Biochem 69(8): 1584-1588. doi: 10.1271/bbb.69.1584
- Zimmer A, Durand C, Loira N, Durrens P, Sherman DJ, Marullo P (2014). QTL dissection of Lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One 9(1): e86298. doi: 10.1371/journal.pone.0086298
- Yuasa N, Nakagawa Y, Hayakawa M, Iimura Y (2004). Distribution of the sulfite resistance gene SSU1-R and the variation in its promoter region in wine yeasts. J Biosci Bioeng 98(5): 394-397. doi: 10.1016/S1389-1723(04)00303-2
- García-Ríos E, Nuévalos M, Barrio E, Puig S, Guillamón JM A new chromosomal rearrangement improves the adaptation of wine yeasts to sulfite. Environ Microbiol 21(5):1771-1781. doi: 10.1111/1462-2920.14586
- Treu L, Toniolo C, Nadai C, Sardu A, Giacomini A, Corich V, Campanaro S (2014). The impact of genomic variability on gene expression in environmental Saccharomyces cerevisiae strains. Environ Microbiol 16(5): 1378-1397. doi: 10.1111/1462-2920.12327
- Park H, Bakalinsky AT (2000). SSU1 mediates sulphite efflux in Saccharomyces cerevisiae. Yeast 16(10): 881-888. doi: Nardi T, Corich V, Giacomini A, Blondin B (2010). A sulphite-inducible form of the sulphite efflux gene SSU1 in a Saccharomyces cerevisiae wine yeast. Microbiology 156(Pt 6): 1686-1696. doi: 10.1099/mic.0.036723-0
- Aranda A, Jimenez-Marti E, Orozco H, Matallana E, Del Olmo M (2006). Sulfur and adenine metabolisms are linked, and both modulate sulfite resistance in wine yeast. J Agric Food Chem 54(16): 5839-5846. doi: 10.1021/jf060851b
- Xu X, Wightman JD, Geller BL, Avram D, Bakalinsky AT (1994). Isolation and characterization of sulfite mutants of Saccharomyces cerevisiae. Curr Genet 25(6): 488-496. doi: 10.1007/bf00351667
- Pilkington BJ, Rose AH (1988). Reactions of Saccharomyces cerevisiae and Zygosaccharomyces bailii to sulphite. J Gen Microbiol 134(10): 2823-2830. doi: 10.1099/00221287-134-10-2823
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11(12): 4241-4257. doi: 10.1091/mbc.11.12.4241
- Ramsdale M, Selway L, Stead D, Walker J, Yin Z, Nicholls SM, Crowe J, Sheils EM, Brown AJ (2008). MNL1 regulates weak acid-induced stress responses of the fungal pathogen Candida albicans. Mol Biol Cell 19(10): 4393-4403. doi: 10.1091/mbc.E07-09-0946
- Fernandes AR, Mira NP, Vargas RC, Canelhas I, Sa-Correia I (2005). Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Biophys Res Commun 337(1): 95-103. doi: 10.1016/j.bbrc.2005.09.010
- Mira NP, Lourenco AB, Fernandes AR, Becker JD, Sa-Correia I (2009). The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res 9(2): 202-216. doi: 10.1111/j.1567-1364.2008.00473.x
- Mira NP, Becker JD, Sa-Correia I (2010). Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 14(5): 587-601. doi: 10.1089/omi.2010.0048
- Simoes T, Mira NP, Fernandes AR, Sa-Correia I (2006). The SPI1 gene, encoding a glycosylphosphatidylinositol-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak-acid food preservatives. Appl Environ Microbiol 72(11): 7168-7175. doi: 10.1128/AEM.01476-06
- Park H, Hwang YS (2008). Genome-wide transcriptional responses to sulfite in Saccharomyces cerevisiae. J Microbiol 46(5): 542-548. doi: 10.1007/s12275-008-0053-y
- Nadai C, Treu L, Campanaro S, Giacomini A, Corich V (2016). Different mechanisms of resistance modulate sulfite tolerance in wine yeasts. Appl Microbiol Biotechnol 100(2): 797-813. doi: 10.1007/s00253-015-7169-x
- Chebaro Y, Lorenz M, Fa A, Zheng R, Gustin M (2017). Adaptation of Candida albicans to Reactive Sulfur Species. Genetics 206(1): 151-162. doi: 10.1534/genetics.116.199679
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, Giaever G (2008). The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320(5874): 362-365. doi: 10.1126/science.1150021
- Schuller C, Mamnun YM, Mollapour M, Krapf G, Schuster M, Bauer BE, Piper PW, Kuchler K (2004). Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol Biol Cell 15(2): 706-720. doi: 10.1091/mbc.e03-05-0322
- Chang EC, Kosman DJ (1990). O2-dependent methionine auxotrophy in Cu,Zn superoxide dismutase-deficient mutants of Saccharomyces cerevisiae. J Bacteriol 172(4): 1840-1845. doi: 10.1128/jb.172.4.1840-1845.1990
- GIRAUD E, IVANOVA A, GORDON CS, WHELAN J, CONSIDINE MJ (2012). Sulphur dioxide evokes a large scale reprogramming of the grape berry transcriptome associated with oxidative signalling and biotic defence responses. Plant Cell Environ 35(2): 405-417. doi: 10.1111/j.1365-3040.2011.02379.x
- Qin G, Wu M, Wang J, Xu Z, Xia J, Sang N (2016). Sulfur Dioxide Contributes to the Cardiac and Mitochondrial Dysfunction in Rats. Toxicol Sci 151(2): 334-346. doi: 10.1093/toxsci/kfw048
- Kurek EJ (1983). An enzymatic complex active in sulphite and thiosulphate oxidation by Rhodotorula sp. Arch Microbiol 134(2): 143-147. doi: 10.1007/bf00407947
- Leichter J, Joslyn MA (1969). Kinetics of thiamin cleavage by sulphite. Biochem J 113(4): 611-615. doi: 10.1042/bj1130611
- Kohl JB, Mellis AT, Schwarz G (2019). Homeostatic impact of sulfite and hydrogen sulfide on cysteine catabolism. Br J Pharmacol 176(4): 554-570. doi: 10.1111/bph.14464
- Teixeira MC, Monteiro PT, Palma M, Costa C, Godinho CP, Pais P, Cavalheiro M, Antunes M, Lemos A, Pedreira T, Sa-Correia I (2018). YEASTRACT: an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res 46(D1): D348-D353. doi: 10.1093/nar/gkx842
- Garde-Cerdán T, Marsellés-Fontanet AR, Arias-Gil M, Martín-Belloso O, Ancín-Azpilicueta C (2007). Influence of SO2 on the consumption of nitrogen compounds through alcoholic fermentation of must sterilized by pulsed electric fields. Food Chem 103(3): 771-777. doi: 10.1016/j.foodchem.2006.09.018
- Ancin-Azpilicueta C, Barriuso-Esteban B, Nieto-Rojo R, Aristizabal-Lopez N (2012). SO(2) protects the amino nitrogen metabolism of Saccharomyces cerevisiae under thermal stress. Microb Biotechnol 5(5): 654-662. doi: 10.1111/j.1751-7915.2012.00343.x
- Kahana-Edwin S, Stark M, Kassir Y (2013). Multiple MAPK cascades regulate the transcription of IME1, the master transcriptional activator of meiosis in Saccharomyces cerevisiae. PLoS One 8(11): e78920. doi: 10.1371/journal.pone.0078920
- Thakur JK, Arthanari H, Yang F, Pan SJ, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, Struhl K, Moye-Rowley WS, Cormack BP, Wagner G, Naar AM (2008). A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452(7187): 604-609. doi: 10.1038/nature06836
- Kim MS, Cho KH, Park KH, Jang J, Hahn JS (2018). Activation of Haa1 and War1 transcription factors by differential binding of weak acid anions in Saccharomyces cerevisiae. Nucleic Acids Res. doi: 10.1093/nar/gky1188
- Jacquet M, Renault G, Lallet S, De Mey J, Goldbeter A (2003). Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J Cell Biol 161(3): 497-505. doi: 10.1083/jcb.200303030
- Gorner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schuller C (2002). Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J 21(1-2): 135-144. doi: 10.1093/emboj/21.1.135
- Durchschlag E, Reiter W, Ammerer G, Schuller C (2004). Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J Biol Chem 279(53): 55425-55432. doi: 10.1074/jbc.M407264200
- Pfanzagl V, Gorner W, Radolf M, Parich A, Schuhmacher R, Strauss J, Reiter W, Schuller C (2018). A constitutive active allele of the transcription factor Msn2 mimicking low PKA activity dictates metabolic remodeling in yeast. Mol Biol Cell 29(23): 2848-2862. doi: 10.1091/mbc.E18-06-0389
- Sadeh A, Baran D, Volokh M, Aharoni A (2012). Conserved motifs in the Msn2-activating domain are important for Msn2-mediated yeast stress response. J Cell Sci 125(Pt 14): 3333-3342. doi: 10.1242/jcs.096446
- Cheng Y, Du Z, Zhu H, Guo X, He X (2016). Protective Effects of Arginine on Saccharomyces cerevisiae Against Ethanol Stress. Sci Rep 6: 31311. doi: 10.1038/srep31311
- VanderSluis B, Hess DC, Pesyna C, Krumholz EW, Syed T, Szappanos B, Nislow C, Papp B, Troyanskaya OG, Myers CL, Caudy AA (2014). Broad metabolic sensitivity profiling of a prototrophic yeast deletion collection. Genome Biol 15(4): R64. doi: 10.1186/gb-2014-15-4-r64
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
This work was funded by INNO-VINE&WINE, Norte-01-0145-FEDER-000038, co-financed by the European Regional Development Fund (ERDF) through Norte 2020 and by ERFD through POCI-COMPETE 2020. Support received by FCT-Portuguese Foundation for Science and Technology (PTDC/EXPL/AGR-TEC/1823/2013 and PTDC/AGR-TEC/3315/2014) and by INTERACT project – “Integrated Research in Environment, Agro-Chain and Technology”, no. NORTE-01-0145-FEDER-000017, in its line of re-search entitled VitalityWine. Support received by Biosys-tems and Integrative Sciences Institute (BioISI; FCT/UID/Multi/04046/2018) and iBB-Institute for Bioen-gineering and Biosciences (UID/BIO/04565/2019) by FCT and from Programa Operacional Regional de Lisboa 2020 (project no. 007317 and PTDC/AGR-TEC/3315/2014_LISBOA-01-0145-FEDER-016834) is also acknowledged. The authors thank Professor Isabel Sá-Correia for the help and guidance in conducting the chemogenomic analysis.
COPYRIGHT
© 2019

Transcriptomic and chemogenomic analyses unveil the essential role of Com2-regulon in response and tolerance of Saccharomyces cerevisiae to stress induced by sulfur dioxide by Lage et al. is licensed under a Creative Commons Attribution 4.0 International License.