Reviews:
Microbial Cell, Vol. 5, No. 3, pp. 119 - 136; doi: 10.15698/mic2018.03.618
pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 ménage-à-trois to stress tolerance and longevity
1 Functional Biology, KU Leuven, Leuven, Belgium.
2 Life and Health Sciences Research Institute (ICVS), School of Medicine, University of Minho, Braga, Portugal; ICVS/3B’s – PT Government Associate Laboratory, Braga/Guimarães, Portugal.
° Contributed equally, in alphabetical order.
Keywords: yeast, ageing, longevity, pH, V-ATPase, Pma1, PKA, TORC1, Sch9.
Abbreviations:
CLS – chronological lifespan
EGOC – EGO complex,
ER – endoplasmic reticulum,
pHc – cytosolic pH,
pHe – extracellular pH,
pHv – vacuolar luminal pH,
RLS – replicative lifespan,
WT - wild type.
Received originally: 29/09/2017 Received in revised form: 24/12/2017
Accepted: 28/12/2017
Published: 12/01/2018
Correspondence:
Joris Winderickx, Functional Biology, KU Leuven, Kasteelpark Arenberg 31 box 2433; Tel: +32 (0)16 321516; joris.winderickx@kuleuven.be
Conflict of interest statement: The authors report no conflict of interest. J.W. declares that he is co-founder and shareholder of the KU Leuven spin-off companies ReMYND nv (Leuven, Belgium) and ADxNeuroSciences nv (Ghent, Belgium), but this did not influence in any way the content of this manuscript, nor is there a link between the studies reported and the activities of the aforementioned companies.
Please cite this article as: Marie-Anne Deprez, Elja Eskes, Tobias Wilms, Paula Ludovico, Joris Winderickx (2018). pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 ménage-à-trois to stress tolerance and longevity. Microbial Cell 5(3): 119-136. doi: 10.15698/mic2018.03.618
Abstract
The plasma membrane H+-ATPase Pma1 and the vacuolar V-ATPase act in close harmony to tightly control pH homeostasis, which is essential for a vast number of physiological processes. As these main two regulators of pH are responsive to the nutritional status of the cell, it seems evident that pH homeostasis acts in conjunction with nutrient-induced signalling pathways. Indeed, both PKA and the TORC1-Sch9 axis influence the proton pumping activity of the V-ATPase and possibly also of Pma1. In addition, it recently became clear that the proton acts as a second messenger to signal glucose availability via the V-ATPase to PKA and TORC1-Sch9. Given the prominent role of nutrient signalling in longevity, it is not surprising that pH homeostasis has been linked to ageing and longevity as well. A first indication is provided by acetic acid, whose uptake by the cell induces toxicity and affects longevity. Secondly, vacuolar acidity has been linked to autophagic processes, including mitophagy. In agreement with this, a decline in vacuolar acidity was shown to induce mitochondrial dysfunction and shorten lifespan. In addition, the asymmetric inheritance of Pma1 has been associated with replicative ageing and this again links to repercussions on vacuolar pH. Taken together, accumulating evidence indicates that pH homeostasis plays a prominent role in the determination of ageing and longevity, thereby providing new perspectives and avenues to explore the underlying molecular mechanisms.
INTRODUCTION
pH homeostasis is of crucial importance for many molecular and physiological processes. In eukaryotic cells, intracellular pH affects protein folding and enzyme activity, is required for vesicle trafficking and impacts organelle function and integrity. As our knowledge of pH homeostasis progresses, it becomes increasingly evident that the proton not only acts as a facilitator for cellular functions but also as a potent second messenger for the regulation of growth and ageing. Dysregulation of pH and lysosomal dysfunction are being linked to numerous human diseases [1], including neurodegenerative disorders [2][3]. Moreover, acidity-dependent probes are used as lysosomal storage disorder-associated markers [4] and acidification of the extracellular space is known to promote cancer metastasis [5]. These observations led to a growing interest in pH-related research and in this domain the unicellular organism Saccharomyces cerevisiae proves to be a valuable model to unravel the fundamental molecular mechanisms underlying pH homeostasis and the relationship between pH, cell growth, stress resistance and longevity.
THE MAIN PLAYERS IN pH HOMEOSTASIS
Cytosolic and organellar pH are tightly controlled in all eukaryotic cells. The two main players of pH homeostasis in yeast are the V-ATPase and Pma1. The V-ATPase is a proton translocating ATPase that pumps protons from the cytosol into the vacuole, endosomes and Golgi compartments. Pma1 is a P-type ATPase that pumps protons over the plasma membrane to acidify the extracellular space. As accurate control of pH is required for pH homeostasis and optimal functionality of the cell and its organelles, it is not surprising that the V-ATPase and Pma1 act in close harmony. Loss of V-ATPase activity leads to a partial mislocalization of Pma1 to the vacuole and other compartments, probably reflecting a compensatory mechanism [6]. In addition to the V-ATPase and Pma1, numerous other proton pumps and exchangers have been identified in yeast, and they probably fine-tune pH control of the cytoplasm and each of the organelles [7]. However, given the importance of pH for organelle-specific functions, surprisingly little is known about their precise regulation and contribution to pH homeostasis.
–
Regulation of the vacuolar V-ATPase
The V-ATPase is a multisubunit enzyme composed of a peripheral V1 sector responsible for ATP hydrolysis, and a membrane-embedded V0 sector responsible for proton translocation [8]. In contrast to mammalian cells, which often exhibit tissue- and/or organelle-specific expression of multiple isoforms of one subunit, yeast cells only have one organelle-specific V0 isoform. Indeed, Vph1-containing V-ATPase complexes are localized at the vacuolar membrane, while Stv1-containing V-ATPase complexes cycle between the Golgi apparatus and endosomes [9]. The activity of Vph1-containing V-ATPases is mainly regulated by reversible disassembly of the V0 and V1 sectors, in which carbon source availability plays a major role. In the presence of glucose, the sectors are assembled at the vacuolar membrane and the V-ATPase is highly active, acidifying the vacuolar lumen. When glucose is scarce, the vacuolar V1 sector dissociates from the V0 sector and the vacuolar luminal pH (pHv) becomes more alkaline [10]. However, this is in no case an all-or-nothing event, as intermediate levels of assembly have been observed with varying nutrient conditions. The exact mechanism by which glucose impacts on vacuolar V-ATPase assembly remains poorly understood though several links with glucose-induced signalling events and glycolysis have been reported as further explained below. Interestingly, the vacuolar V-ATPase is also responsive to changes in cytosolic (pHc) and extracellular pH (pHe) [11][12][13]. For instance, V-ATPase disassembly upon glucose-starvation is significantly reduced when cells are grown at pH 7 and accordingly, enhanced V-ATPase activity is observed in isolated vacuoles from cells grown under these conditions as compared to cells grown at pH 5 [11][12]. Hence, the V-ATPase appears to act as pH sensor and both in yeast and mammalian cells the vacuole-specific V0 subunit ‘a’, encoded by VPH1 in yeast, has been proposed as pH-sensing protein [11][14][15]. Interestingly, this subunit also mediates the dissociation of the V1 and V0 complexes upon glucose-starvation [11]. In line with this, V-ATPases that reside in the Golgi compartment, containing the STV1-encoded subunit ‘a’ instead of Vph1, do not dissociate upon glucose depletion [16]. Besides glucose and pH, lipids also impact on V-ATPase regulation as the signalling phosphoinositides PI[3][5]P2 and PI[4]P promote V-ATPase activity at the vacuole and Golgi, respectively, through interaction with the appropriate ‘a’ subunits Vph1 and Stv1 [17][18][19]. Additional levels of V-ATPase regulation include the adjustment of coupling efficiency between ATP hydrolysis and proton translocation, and the mechanisms for assembly of the V0 complex in the endoplasmic reticulum (ER) and the subsequent export and delivery of the vacuolar V-ATPase to the vacuolar membrane [20][21][/cite][22][23][24].
–
Regulation of the plasma membrane embedded Pma1
The Pma1 P-type ATPase is considered as the major determinant of pHc and plasma membrane potential in yeast [25][26]. In contrast to genes encoding subunits of the V-ATPase, PMA1 is an essential gene, making the protein a difficult target to study. Different environmental and nutritional factors control Pma1 activity, with glucose availability being the best studied. The transcription of PMA1 is enhanced during growth on glucose [27]. In addition, the proton pump is controlled by reversible phosphorylation, which modulates an inhibitory interaction of the C-terminus with the active site of the H+-ATPase. The latter reflects a complex interplay of several signalling events, albeit not all players have yet been identified. Evidence obtained so far points to an involvement of the casein kinase 1 homologues Yck1 and Yck2, the protein kinase Ptk2 and the protein phosphatase Glc7 [28][29][30]. Calcium signalling was also shown to modulate the plasma membrane H+-ATPase activation in response to glucose [31]. Furthermore, the intra- and extracellular pH, as well as the plasma membrane potential affect Pma1 activity. Here, an important contribution has been ascribed to the pH-dependent regulation of the potassium transporter Trk1 and the compensatory roles of K+ transport and H+ efflux to maintain the electrochemical gradient [6][32]. As mentioned before, a reduction in V-ATPase activity, for instance by glucose starvation, triggers sorting of Pma1 from the Golgi to the vacuole [33].
–
Note that the yeast genome encodes for another plasma membrane H+-ATPase, i.e. Pma2, but the expression of this pump is very low and, consistently, it only has a minor impact on cellular pH [34][35].
–
The proton gradients established by the V-ATPase, Pma1 and other proton transport systems are of absolute importance for several processes. For instance, the driving force created by these gradients is crucial to maintain phosphate homeostasis [36] and the cation balance of alkali metals (Na+ and K+), divalent cations (Ca2+ and Mg2+) and trace metals (Fe2+, Zn2+, Cu2+ and Mn2+). Their symport/antiport along with H+ helps to maintain their physiological and non-toxic levels, so as to provide a suitable environment for various biochemical reactions. The exact regulation of this cation balance has been the subject of an excellent review [7]. Several membrane proteins rely on the proton driving force. Examples are the proton-coupled phosphate symporters and the different amino acid permeases that reside in the plasma membrane [36][37] or the yeast AVT1-7 family members that mediate bidirectional transport of amino acids across the vacuolar membrane [38].
A MÉNAGE-À-TROIS FOR NUTRIENT SIGNALLING AND pH REGULATION
As both the V-ATPase and Pma1 react closely to nutrient availability, it seems evident that proton pumping activity and pH homeostasis must be regulated by an interplay of diverse nutrient-induced signalling networks. Indeed, the protein kinases PKA and Sch9 and the protein kinase complex TORC1 play a central role. This so-called ménage-à-trois integrates input from several nutrient sensing systems in order to regulate metabolism, intracellular trafficking, proteome integrity, autophagy, stress resistance, cell size, cell cycle progression, growth and sporulation [39][40][41][42]. The activity of each of these kinases is tightly regulated in response to different nutritional cues.
–
Nutrient-controlled regulation of the ménage-à-trois
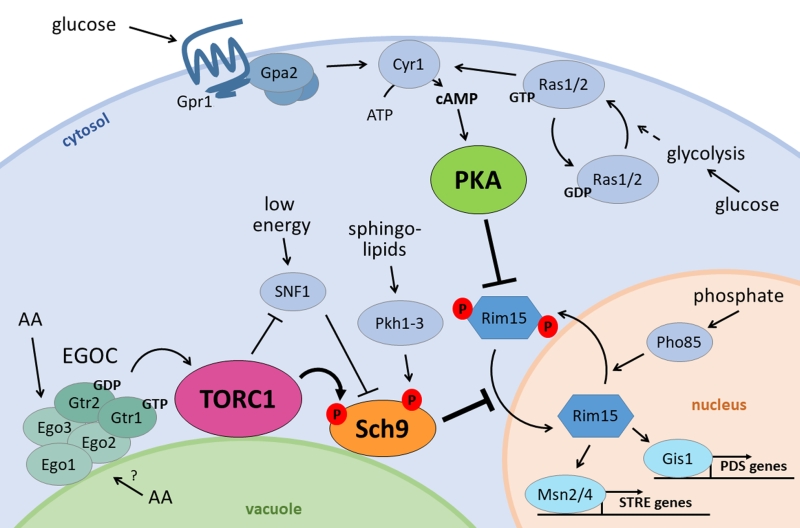
As shown in Fig. 1, the activity of PKA is regulated by the Ras-cAMP pathway and activation of adenylate cyclase, the latter depending on extracellular glucose sensing via the Gpr1/Gpa2 GPCR system as well as on intracellular glucose sensing via activation of the small G-proteins Ras1 and Ras2 [43]. The protein kinase complex TORC1 is regulated by intracellular amino acid signalling and this occurs by different mechanisms depending on the quality of the amino acid as a nitrogen source [44]. Leucine and probably other neutral amino acids signal to TORC1 through the EGO complex (EGOC), the orthologue of the mammalian Rag-Ragulator complex{Powis, 2016 #279}{Powis, 2016 #279}. EGOC consists of Ego1, Ego2 and Ego3 that form a scaffold at the vacuolar membrane for the Rag GTPases Gtr1-2. The latter function as a heterodimer that receives input about the cytosolic amino acid content. When amino-acids are available in the cytoplasm, an active EGOC formation is triggered in which Gtr1 is in its GTP-bound form and Gtr2 in its GDP-bound form, leading to TORC1 activation. Amino acid starvation induces the opposite GTP/GDP-loading status of the GTPases and the consequent inhibition of TORC1 [45][46]. Recent evidence obtained in mammalian systems suggests a role for the Rag-Ragulator in signalling lysosomal amino acid content and transport in addition to cytosolic amino acid content [47][48], but whether EGOC has a similar role in sensing the vacuolar amino acid load in yeast remains to be studied. Preferred nitrogen sources like glutamine can signal independently of EGOC to trigger a more sustained TORC1 activation required to support vigorous growth [44]. At present, the underlying mechanism for this Gtr1/2-independent TORC1 activation in yeast remains poorly understood, though one study identified Ypt1, a GTPase involved in ER-to-Golgi vesicular trafficking, as an alternative regulator of TORC1 [49]. Other studies suggested a role for the vacuolar membrane-associated phosphatidylinositol (PI) 3-phosphate binding protein Pib2 and the Vps15/34 Pl 3-kinase complex [50][51]. The third member of the ménage-à-trois, the AGC kinase Sch9, is the yeast orthologue of mammalian PKB/Akt and S6 kinase. It is a well-known target of TORC1 and as such it is involved in amino acid and nitrogen source signalling [52]. However, Sch9 has been implicated in the sensing and signalling of other nutrients as well. It plays a major role in lipid signalling as a regulator of the so-called sphingolipid rheostat [53] and, besides its phosphorylation by TORC1, Sch9 needs to be phosphorylated by sphingolipid-activated kinases Pkh1-3 at its PDK1 site in the activation loop to obtain full activity [52][54][55]. Moreover, Sch9 also responds to glucose availability, thereby acting in conjunction with PKA [56], and receives input of the protein kinase complex SNF1, a central regulator of cellular energy homeostasis [57]. Thus, Sch9 appears to play an integratory role in nutrient sensing that allows for coordination and fine-tuning of diverse signalling cascades. As such, it shares several effectors with the PKA and TORC1 signalling routes but for several of these targets the effect inflicted by Sch9 can either be additive or opposite depending on the activity status of the other kinases [41][58][59][60].
–
An example of the interplay between the signalling pathways controlled by the ménage-a-trois is their convergence to regulate the activity and nuclear localization of Rim15, a protein kinase required for metabolic adaptation and the general stress response (Fig. 1) [61]. Here, also phosphate availability comes into the picture as the nuclear exit of Rim15 is controlled by the phosphate-responsive PHO-pathway, particularly Pho85 [41][62].
–
The nutrient status regulates pH homeostasis
The most straightforward evidence for the role of nutrient signalling in pH regulation are the alterations of extra- and intracellular pH in response to nutrient availability and growth rate. Albeit quite complex, we can summarize this as follows. When cells grow fermentatively, they produce organic acids that acidify the medium, such as acetic acid. In its protonated form, acetic acid is able to move through the plasma membrane back into the cell [63]. Once inside the cytosol, whose pH is maintained around neutrality during exponential growth, protons dissociate from the weak acid causing intracellular acidification. This acidification is counteracted by Pma1-mediated proton efflux and V-ATPase-mediated vacuolar acidification in order to maintain pHc homeostasis [9][64][65][66]. During the diauxic shift or upon glucose starvation, the activity of Pma1 reduces, the cytosol acidifies and the vacuolar V-ATPase disassembles [11][67][68][69][70]. Re-addition of glucose or sucrose to glucose-starved cells initially results in a rapid drop in pHc, probably due to the resumption of glycolysis, but this is quickly followed by intracellular alkalization and extracellular acidification as a result of Pma1 and vacuolar V-ATPases regaining full activity [11][70][71][72]. In contrast to the clear impact of carbon source availability, re-addition of amino acids or another nitrogen source to starved cells does not affect pHc [70][73]. This demonstrates that adjusting pHc is not simply a matter of growth resumption, but that it is specifically controlled by carbon source availability.
–
The ménage-à-trois is crucial for pH regulation
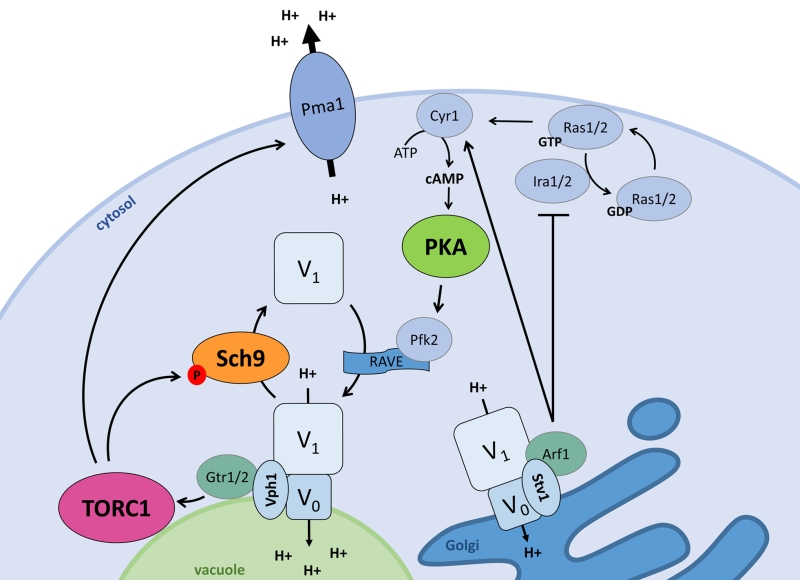
The ménage-à-trois plays an important role to integrate nutrient availability and pH homeostasis as shown in Fig. 2. Alterations that enhance PKA activity, like deletion of the Ras2 GTPase IRA2 or the PKA regulatory subunit BCY1, were found to inhibit vacuolar V-ATPase disassembly upon glucose deprivation. This suggests that PKA is involved in glucose-dependent V-ATPase regulation [74]. Although PKA-dependent phosphorylation has been described for the V1 subunit ‘C’ in insect cells [75] and for the V1 subunit ‘A’ in human HEK-293T cells [76], no such event has been reported in yeast so far. Nonetheless, the Ras-cAMP pathway and PKA are known to regulate key enzymes of glycolysis in yeast [41][77] and two glycolytic enzymes, i.e. aldolase and phosphofructokinase, were reported to associate with the V-ATPase, placing them in pole position to mediate the glucose signal [67][78][79][80]. Especially the phosphofructokinase Pfk2 might be a good candidate as it is a direct target of PKA [81]. Pfk2 is indeed required to maintain vacuolar acidification and optimal RAVE-mediated reassembly of the V0 and V1 subunits upon re-addition of glucose to cells that were briefly deprived of the sugar [67][78]. RAVE is the acronym for ‘Regulator of H+-ATPase of Vacuolar and Endosomal membranes’ and is a complex known to function as a scaffold that binds the V0 and V1 sectors in a glucose-dependent manner [82][83]. Earlier studies also suggested a possible role for the Ras-cAMP pathway in the glucose-induced activation of Pma1 [84][85]. However, subsequent detailed analysis contradicted such a role and showed that the plasma membrane H+-ATPase is still activated in mutant strains deficient for glucose-induced cAMP increase and that similar activation levels are obtained in strains with normal or attenuated PKA activity [86][87].
–
Although starvation and resupplementation of nitrogen and amino acids does not influence pHc [70][73], both TORC1 and Sch9 are involved in the regulation of pH homeostasis in function of glucose availability. When cells are grown on glucose both players are localized at the vacuolar membrane, but once cells enter the diauxic shift, Sch9 is displaced into the cytosol. The latter occurs concomitantly with the disassembly of the vacuolar V-ATPase [70]. In fact, Sch9 has a modulatory role in controlling the V-ATPase assembly state as evidenced by the observation that cells lacking Sch9 display an enhanced association of the V0 and V1 sectors in the presence of glucose and a delayed disassembly of the proton pump upon glucose starvation. Similar effects were also seen upon rapamycin-induced TORC1 inhibition or upon replacement of the wild type Sch9 allele by the Sch95A version that can no longer be activated by TORC1 [70]. Accordingly, sch9D cells have more acidic vacuoles both in the presence or absence of glucose, and while sch9D mutant cells are able to maintain their pHc within the same range as wild type (WT) cells during fermentative growth, once they traverse the diauxic shift a hyperacidification of the cytosol is observed [70]. Whether the latter points to a role of Sch9 in the regulation of Pma1 or other ion pumps and exchangers remains to be investigated in more detail, but at least two observations suggest that this may well be the case. Indeed, sch9D cells fail to rapidly acidify the extracellular medium when glucose is refed to glucose-starved cells and the additional deletion of SCH9 in strains lacking a functional V-ATPase triggers a further alkalization of the vacuolar lumen [70]. Interestingly, a most recent paper reported that TORC1 is required to obtain full Pma1 activity and although a possible role of Sch9 was not examined, the study demonstrated an involvement of the protein phosphatase Sit4, another TORC1 effector, in the turnover but not the phosphorylation status of Pma1. The same study also showed that both sit4D cells and cells lacking the TORC1 subunit Tco89 display reduced K+ uptake and a more acidic intracellular pH as compared to WT cells, but solely in the sit4D mutant this was associated with a reduced Pma1 activity [88].
–
Additional players link nutrient signalling with pH homeostasis
Apart from the effects mediated by the PKA and TORC1-Sch9 axis on V-ATPase assembly/disassembly or Pma1 activity, other players involved in nutrient signalling also affect pH homeostasis. As mentioned, the signalling lipid PI[3][5]P2 was shown to be important for pHv regulation. Accordingly, the PI[3][5]P2-deficient mutants vac7, vac14 and fab1 all display vacuolar acidification defects [89][90][91]. Although PI[3][5]P2 may directly affect V-ATPase activity [18], the phosphoinositide may also act indirectly via TORC1/Sch9 signalling, as TORC1 activity as well as the recruitment of Sch9 and its TORC1-dependent phosphorylation at the vacuolar membrane are dependent on the presence of PI[3][5]P2 [92][93][94]. Moreover, increased PI[3][5]P2 levels are associated with vacuolar fragmentation, which involves TORC1 and its downstream effectors [95][96]. This relationship is further highlighted by the significant overlap in targets between a genome-wide screening aiming to identify genes conferring a synthetic sick/lethal phenotype when combined with the SCH9 deletion and a screening for genes mediating inositol auxotrophy [70][97]. Similar genome-wide screenings were performed to find proteins involved in the control of pHc or pHv during fermentative growth on glucose [24][25] and at least with the screening on pHc a significant overlap is seen when compared to the data of the synthetic SCH9 screening [70]. Besides proteins associated with vesicular transport, lipid metabolism or amino acid biosynthesis, both pH screenings retrieved proteins required formitochondrial functions. This is intriguing because it indicates that mitochondria are essential to maintain pH homeostasis even during fermentative growth, where they are not required for energy supply. This underscores the importance of cross-talk between organelles, and the notable lack of research performed to date in this area.
–
pH as a second messenger for nutrient signalling
Changes in intracellular pH affect cell functioning at different levels as it impacts on protein folding, enzyme activities and the protonation of biological macromolecules, lipids and other metabolites [66]. The crucial question, however, is whether alterations in pH are sensed and perceived as signals. Perhaps the best way to answer this is to describe the recent advances in how pH affects the activities of the PKA/TORC1/Sch9 ménage-à-trois.
–
Already decades ago, it was reported that the treatment of fungi with depolarizing agents inflict a rapid increase in the level of cAMP [98][99][100]. Mechanistically, different scenarios were described as intracellular acidification was found to enhance both the affinity of the adenylate cyclase Cyr1 for its substrate ATP [98] as well as Ras-GTP loading by inhibition of the GTPase-activating proteins, Ira1 and Ira2 [101]. Notably, a mild treatment of cells with a protonophore allows to bypass the requirement of hexose transport for glucose-induced activation of the Ras-cAMP pathway [102], which is consistent with the observation that the protonophore treatment of glucose-starved cells triggers a rapid drop in pHc similar as that seen immediately after addition of glucose to these starved cells [98]. A more recent study correlated the glucose-induced changes in pHc to V-ATPase assembly/disassembly and subsequent activation of PKA [11]. As mentioned, this study also proposed the V‑ATPase to act as pH-sensor with Vph1, the V0 subunit ‘a’, as putative pH-sensing protein. Subsequently, the role of the V-ATPase for activation of the Ras-cAMP pathway was further defined, since in the presence of glucose the pump was shown to signal to the Ras proteins via Arf1, a GTPase that interacts with the Golgi-specific V0 subunit Stv1. Moreover, the activity of the plasma membrane ATPase Pma1 was shown to stimulate growth through Ras activation by increasing pHc [73].
–
Additionally, pHc and the V-ATPase also confer signals for glucose availability to TORC1 and Sch9. To this end, the V-ATPase controls the GTP-load of Gtr1 and interacts with this EGOC Rag GTPase via the vacuole-specific V0 subunit Vph1. This finding is rather remarkable as Gtr1 plays a pivotal role in amino acid sensing [45][46], but amino acid availability itself does not affect pHc nor the V-ATPase assembly state [70][73]. As the V-ATPase apparently acts as activator of both PKA and the TORC1-Sch9 axis, while each of the kinases has the ability to provide feedback by affecting the V-ATPase assembly/disassembly, a very balanced system based on feedback loops is established (Fig. 2). The necessity of this tight control relates undoubtedly to the crucial roles of the PKA/TORC1/Sch9 ménage-à-trois for the overall cellular functioning and the coordination between growth and the cell cycle [41]. The latter is further evidenced by the finding that the TORC1-Sch9 axis transmits signals from the vacuole that are required for cell cycle progression [94].
–
A signalling role of pHc also became apparent from data obtained from a genome-wide screening that investigated the correlation between aberrant intracellular pH and reduced growth rate of mutants. This analysis confirmed a tight connection between both, suggesting that pHc dictates the growth rate. For 19 out of the 173 mutants, however, the causal relationship between pHc and growth rate was completely abrogated, indicative that these mutants fail to properly sense the pH signal. Among them were mutants affected in mitochondrial translation, inositol phosphate biosynthesis and lipid biosynthesis [25]. Importantly, a similar study found no causal correlation between pHv and growth rate [24].
pH CONTROL, STRESS TOLERANCE AND LONGEVITY
It is well established that the nutrient pathways controlled by the PKA/TORC1/Sch9 ménage-à-trois have a significant impact on cellular ageing, whether being monitored as replicative lifespan (RLS) by assessing the number of divisions a mother cell can undergo before dying, or as chronological lifespan (CLS) by assessing the time span a non-dividing cell remains viable [103]. Both modes of ageing rely on partially overlapping cellular and molecular determinants and these have been the topic of excellent reviews to which we refer for more details [40][104][105][106][107][108][109][110][111][112]. As nutrient availability and the PKA/TORC1/Sch9 ménage-à-trois affect pH control, it is not surprising that pH is being linked to the regulation of ageing/longevity in yeast. Hence, a number of groups have explored the interplay between extracellular, cytosolic and organelle pH and longevity, the topic of the sections below and summarized in Fig. 3.
–
Acetic acid shortens longevity and induces programmed cell death
A nice example that illustrates the interconnections between lifespan, nutrient availability and their dependence on pH is the observation that chronologically aged cells display a reduced subsequent RLS, but that this can be attenuated when the chronological ageing occurs in buffered medium or in calorie restriction (CR) conditions [113][114]. As mentioned, during fermentative growth yeast cells produce and secrete different organic acids. While some of these acids accumulate in the growth medium, acetic acid levels decline after post-diauxic shift, indicating that this acid is used as a carbon source. The drawback is, however, that acetic acid uptake by the cell comes with a degree of toxicity that affects longevity (Fig. 3). Consistently, growing cells under acidic conditions was found to shorten CLS and RLS, while CR significantly reduces the production of organic acids and extends CLS and RLS [114][115][116]. Buffering the medium also has a positive effect on lifespan as it lowers the difference between the extra- and intracellular pH values, thereby reducing the driving force for inwards diffusion of acetic acid. As such, buffering presumably prevents stress by reducing the amount of energy that needs to be consumed to maintain the intracellular pH [117].
–
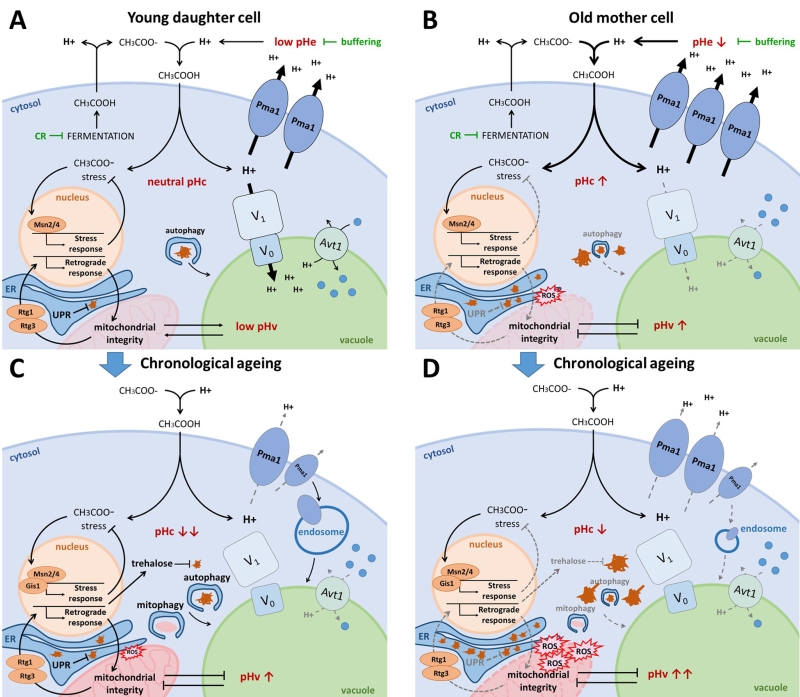
Indeed, as a first line of defense against acetic-acid induced stress, cells rely on energy-consuming proton pumps to counteract cytosolic acidification caused by dissociation of protons from the acetic acid once it has entered the cell, and to maintain pH optima in all different cellular compartments. Here, the plasma membrane-embedded Pma1 fulfils an important role by extruding these protons from the cytosol, allowing healthy exponential WT cells to maintain pHc around neutrality independently of pHe (Fig. 3A) [71]. During ageing however, yeast cells struggle to maintain proper pH homeostasis. As mother cells undergo divisions, the pHc increases whereas daughter cells retain a more acidic cytosol (Fig. 3B). This phenomenon is attributed to asymmetrical distribution of Pma1, which predominantly remains in the plasma membrane of the mother cell [118]. This mother-specific increase in Pma1 activity and subsequent reduction of cytosolic proton content is believed to trigger the decline of vacuolar acidity during replicative ageing, as protons are unavailable for the V-ATPase to be pumped into the vacuolar lumen [118]. Conversely, when cells have traversed the diauxic shift or when encountering glucose depletion, a condition typically used to study chronological ageing, the activity of Pma1 declines as mentioned previously. Moreover, also the V-ATPase is disassembled under these conditions [11][67][68][69][70] and this loss of V-ATPase activity signals ubiquitination and endocytosis of Pma1 [119]. In consequence, the pHc is significantly reduced both in post-diauxic cells and in chronologically aged cells and due to the lower activity of the V-ATPase, the vacuole presumably becomes more alkaline (Fig. 3C).
–
Secondly, mild stress induces the so-called general or environmental stress response pathway, an adaptive response that allows to acquire a form of stress resistance that protects cells from subsequent stress triggered by the same or another stressor [120][121]. This adaptive response applies to acetic acid-induced stress as well as to physiological stresses affecting lifespan [122] and it is the basis of hormesis effects that play in ageing [123][124]. In the environmental stress response pathway, the transcription factors Msn2/4 play a central role. Msn2/4 are well known targets of the PKA/TORC1/Sch9 ménage-à-trois required to induce expression of several stress-responsive genes needed to protect cells from adverse conditions (Fig. 1; Fig. 3) [41]. Notably, these transcription factors are also induced when cells encounter alkaline stress [125][126], suggesting that the actual stress factor might be a change in pH, the proton gradient or membrane potential. A study that aimed to identify genes essential for the acquisition of tolerance to different weak acids implicated Msn2/4 for acetic acid tolerance [127]. Interestingly, apart from vacuolar acidification, intracellular trafficking and ergosterol biosynthesis, acetic acid tolerance was found to specifically depend on a small set of genes and this included those encoding Ras2, the trehalose-6P synthase, different cytosolic and mitochondrial ribosomal proteins and two well-known players of the retrograde response pathway, i.e. Rtg2 and Rtg3 [127]. These data nicely complement observations connecting metabolism, cell death and longevity. The finding of mitochondrial ribosomal proteins and components of the retrograde response pathway is consistent with the requirement of functional mitochondria to maintain pH homeostasis [24][25], and there is ample evidence that mitochondrial dysfunction accompanies acetic acid-induced programmed cell death (PCD) [128][129] and the reduction of CLS and RLS (Fig. 3B-D) [130][131][132][133][134][135]. Upon mitochondrial dysfunction, the retrograde response pathway is triggered to transmit signals to the nucleus in order to make adjustments in cellular metabolic and biosynthetic activities. The retrograde response pathway is positively controlled by Ras2, explaining why this small GTPase is essential for the acquisition of acetic acid tolerance. In fact, PKA, similar as TORC1, negatively influences the retrograde response pathway [136]. Among other genes, the retrograde response pathway targets several cytosolic and mitochondrial ribosomal protein genes [137], explaining their involvement in acetic acid tolerance.
–
Some evidence suggests that the expression of genes involved in trehalose biosynthesis is also influenced by the retrograde response pathway [137]. Although the role of trehalose in conveying acetic acid tolerance is not fully understood, the long-lived tor1D and sch9D mutants appear to make optimal use of the protective properties of trehalose as these strains were found to switch their metabolism in the quiescent phase and use the acetic acid that was secreted during the pre-diauxic phase to produce trehalose [138]. A recent study reported that Tps1 can decelerate chronological ageing independently of its known trehalose-6P catalytic activity [139][140]. Consistently, the tps1D mutant is short-lived. However, another study demonstrated that this phenotype is shared by mutants lacking the trehalose-6P phosphatase Tps2, or the neutral trehalases Nth1 and Nth2. In contrast, mutants lacking the regulatory subunits of the trehalose synthase complex Tps3 or Tsl1, or the periplasmic acid trehalase Ath1, were found to be long-lived [141]. Analysis of these different mutants revealed that trehalose reduces the amount of oxidative carbonylated proteins during post-diauxic phase and that it lowers the level of protein aggregation in the quiescent state [141]. Thus, trehalose seems to protect cells by preventing proteotoxicity (Fig. 3C-D).
–
pH and proteotoxicity
Cells are equipped with a complex network that ensures proteome integrity. Proteins that can no longer fulfil their function due to misfolding, damage or aggregation become substrates for the protein degradation machinery and they are cleared either via the proteasome, or via autophagy and vacuolar targeting (Fig. 3). Here the link to pH homeostasis is obvious, as the final step of autophagy, i.e. the disintegration of autophagic bodies, is linked to vacuolar membrane integrity and acidification of the vacuolar lumen by the V-ATPase [142][143][144]. Next to its detoxifying function, however, autophagy also has a role in nutrient recycling. In that regard, it seems logical that autophagy is upregulated in nutrient limiting conditions and that the nutrient signalling ménage-à-trois plays an important role in the regulation of autophagy [40][41][60]. Interestingly, the cellular capacity for autophagic degradation declines with age, which will itself also contribute to the accumulation of cellular and molecular damage. Accordingly, induction of autophagy extends lifespan, and this seems to be accompanied by vacuolar acidification (Fig. 3) [143][145].
–
Recent evidence indicates that misfolded and damaged proteins are first partitioned in specific inclusions in the cell. In JunQ and INQ misfolded proteins are refolded, while those deposited in IPOD probably await clearance via autophagy [146][147]. This partitioning requires Hsp104 and functional actin cables [148][149][150], the latter being another essential lifespan determinant [151][152]. Partitioning of misfolded proteins is beneficial during chronological ageing [153] and assures the asymmetric inheritance of damaged and non-functional proteins during replicative ageing, thereby producing rejuvenated daughter cells [148][149][150]. A recent study connected the V-ATPase, vesicular trafficking and components involved in actin cable-dependent vacuole inheritance to this asymmetric inheritance [154], implying a pH dependency. Notably, apart from the vacuole also the ER, mitochondria and even mRNA are subject to asymmetric inheritance during the division of yeast cells (reviewed in [155]).
–
Mitochondrial dysfunction and its interplay with the ER and the vacuole
Several molecular chaperones assist in the correct folding of proteins or the refolding in case protein misfolding occurs. These chaperone proteins can be found in the cytoplasm, the ER and mitochondria. In fact, a recent study revealed that yeast cells possess a cell-wide proteostasis system where proteotoxicity in one cellular compartment triggers a response in other compartments. This response, termed cross-organelle stress response (CORE) has a protective role and extends both CLS and RLS [156]. Aberrant chaperone activity in each of the compartments leads to fragmented mitochondria, a loss of respiratory activity and an increase in cytosolic NADPH reducing power. This effect is associated with inactivation of TORC1, which acts as protein folding sensor, and the subsequent activation of Snf1 [156][157]. The existence of a cell-wide proteostasis system is also inferred by the observation that yeast cells use a common system to monitor and ensure protein quality control in the ER and mitochondria. This system involves Cdc48/p97, an AAA ATPase known from the ubiquitin-proteasome system that is recruited by stressed ER or mitochondria to extract ubiquitinated proteins presented at the membranes of the organelles and to direct these to the proteasome [158][159]. Interestingly, several physical organelle contact sites exist in yeast, although their involvement in CORE and the cell-wide proteostasis system remains to be elucidated. Here we mention ERMES, the ER-mitochondrial contact site [160], and vCLAMP, the vacuole-mitochondrion contact site [161]. Several nutrient and ion transporters have been shown to be enriched at these contact sites, indicating a role as hubs for the exchange of nutrients and ions between organelles [161] and pointing towards the idea that pH could be an essential regulator. In addition, ERMES and vCLAMP have been proposed to serve as dynamic metabolic signalling hubs [162]. Notably, they are co-regulated in response to nutrients and appear to fulfil partially overlapping functions as their simultaneous disruption is lethal [161].
–
It is obvious that mitochondria play an important role in the determination of lifespan that surpasses their function as energy supplying factory. It is essential for viability that dysfunctional mitochondria are removed, which occurs via mitophagy [40]. Efficient mitophagy requires ubiquitination of the ERMES components Mdm34 and Mdm12 [163]. Moreover, ERMES colocalizes with the site of mitophagosome generation and the ER was proposed to deliver the necessary lipids for membrane engulfment of the mitochondrion [164][165]. Whether vCLAMP has a role in mitophagy has not been investigated yet. Nonetheless, it is known that it suffices to enhance vacuolar proton pumping to significantly reduce mitochondrial dysfunction, indicating a communication between these organelles which could rely on vCLAMP. Consistently, cells lacking functional V-ATPases display significant increased levels of markers for dysfunctional mitochondria and a dramatically shortened RLS and CLS (Fig. 3B-D) [70][134]. Intriguingly, this appears not to be related to a reduced capacity of vacuolar protein degradation, but rather to the impaired ability of the vacuole to efficiently store amino acids. This is evidenced by the observation that enhanced expression of the neutral amino acid transporter Avt1 attenuates mitochondrial dysfunction in replicative ageing cells without preventing vacuolar alkalization [134]. How this relates to the regulation of autophagic processes by the PKA/TORC1/Sch9 ménage-à-trois [40][166][167] remains to be investigated.
–
pH, lipid synthesis and liponecrosis
The synthesis of major membrane lipids is spatially organized and involves different organelles [168]. Moreover, many lipid synthesizing enzymes are enriched at the contact sites between the ER and other organelles and these contacts mediate non-vesicular selective lipid transport [169][170][171][172][173][174][175]. As organelles rely on pH for optimal functionality, it is expected that compartmental cross-talk for lipid metabolism is closely connected to pH regulation. This is supported by the observation that membrane contact sites constitute domains important for ion transport as well [161][176], which in case of the ER-plasma membrane contact is linked to proton pumping by Pma1 [177]. Moreover, a study that used a systems biology approach to investigate the interdependence of pH control and CLS by comparison of young and old cells grown in buffered or non-buffered medium revealed that pH has a main impact on the reorganization of lipid metabolism. This reorganization has a beneficial effect on CLS by preserving mitochondrial and vacuolar health, the latter being dependent on V-ATPase activity [178].
–
Lipid homeostasis is of utmost importance to maintain longevity of yeast cells and persistent deviations thereof can lead to cell demise and a phenomenon described as liponecrosis [105][179][180][181]. Several observations indicate that alterations in lipid homeostasis induce mechanisms involved in protein quality control. Conditions that perturb lipid biosynthesis, that alter the lipid compositions of the plasma membrane and endomembranes or that affect the lipid droplet content of cells were all reported to trigger ER stress and activation of the unfolded protein response (UPR) [182][183][184][185][186][187][188]. In contrast to acute ER stress, which induces PCD [189][190], the lipid-associated induction of ER stress appears to be linked to compensatory mechanisms directed to reinstate lipid biosynthesis and lipid metabolism and to promote cell survival [191][192][193][194][195]. For instance, the activation of the UPR was shown to restore normal ceramide levels when sphingolipid biosynthesis was compromised [192] and to enhance synthesis of triacylglycerols and sterol esters in order to stimulate the formation of lipid droplets [196]. Besides their role for energy storage, these lipid droplets are essential for the regulation of autophagic processes and the clearance of damaged and aggregated proteins from the ER and mitochondria [197][198][199][200][201]. Interventions that affect lipid homeostasis are commonly associated with the appearance of fragmented vacuoles and V-ATPase dysfunction. This includes alterations in the biosynthesis of ergosterols, sphingolipids and ceramides or the availability of essential precursors like inositol [202][203][204][205]. However, preventing lipid droplet formation by blocking the synthesis of di- and triacylglycerol through deletion of the PAH1-encoded phosphatidic acid phosphatase appears to be an exception since pah1D cells are characterized by vacuolar fragmentation and enhanced lipid toxicity while still displaying improved acidification of the vacuolar lumen due to elevated expression of V-ATPase subunits [206][207]. The latter relates to a negative effect of Pah1 on the transcription of several V-ATPase subunit genes, which all contain an UASINO element in their promotor [206]. This is interesting since this promotor element also links the expression of these V-ATPase genes to phospholipid biosynthesis and the availability of inositol, choline and phosphate [208].
CONCLUDING REMARKS
Taken together, control of pH homeostasis is emerging as a key factor determining longevity and alterations culminate in many hallmarks of ageing. Although we are only beginning to uncover the importance of pH homeostasis, it is already amazing how many aspects of cell functioning are influenced by intracellular pH and the reciprocal regulation of mainly two players, the V-ATPase and Pma1. When thriving in an environment with plentiful nutrients, Pma1 and the V-ATPase maintain an ideal pH to support vigorous cell growth. During ageing, the activity of these two players changes, and while increased Pma1 activity may provide enhanced tolerance of a mother cell to the weak acids that were produced, the drawback is a reduction of vacuolar acidity. Indeed, vacuolar acidity is of absolute importance for endocytosis and vesicular trafficking, as well as cellular damage control, the latter via the clearance of malfunctioning proteins and organelles, important for both CLS and RLS (Fig. 3). In addition, damage control also occurs via asymmetrical inheritance during replicative ageing and also here links with vacuolar functioning and pHv are emerging. Undoubtedly, this field of research opens a promising path towards the understanding of intrinsic mechanisms of ageing and longevity, which could be of critical value for our insight into lysosomal-related human diseases and proteopathies, including neurodegenerative disorders, different types of cancer and lysosomal storage diseases.
References
- F.M. Platt, "Sphingolipid lysosomal storage disorders", Nature, vol. 510, pp. 68-75, 2014. http://dx.doi.org/10.1038/nature13476
- M.B. Bagh, S. Peng, G. Chandra, Z. Zhang, S.P. Singh, N. Pattabiraman, A. Liu, and A.B. Mukherjee, "Misrouting of v-ATPase subunit V0a1 dysregulates lysosomal acidification in a neurodegenerative lysosomal storage disease model", Nature Communications, vol. 8, 2017. http://dx.doi.org/10.1038/ncomms14612
- R.U. Onyenwoke, and J.E. Brenman, "Lysosomal Storage Diseases-Regulating Neurodegeneration", Journal of Experimental Neuroscience, vol. 9s2, pp. JEN.S25475, 2015. http://dx.doi.org/10.4137/jen.s25475
- D. te Vruchte, A.O. Speak, K.L. Wallom, N. Al Eisa, D.A. Smith, C.J. Hendriksz, L. Simmons, R.H. Lachmann, A. Cousins, R. Hartung, E. Mengel, H. Runz, M. Beck, Y. Amraoui, J. Imrie, E. Jacklin, K. Riddick, N.M. Yanjanin, C.A. Wassif, A. Rolfs, F. Rimmele, N. Wright, C. Taylor, U. Ramaswami, T.M. Cox, C. Hastings, X. Jiang, R. Sidhu, D.S. Ory, B. Arias, M. Jeyakumar, D.J. Sillence, J.E. Wraith, F.D. Porter, M. Cortina-Borja, and F.M. Platt, "Relative acidic compartment volume as a lysosomal storage disorder–associated biomarker", Journal of Clinical Investigation, vol. 124, pp. 1320-1328, 2014. http://dx.doi.org/10.1172/jci72835
- Y. Kato, S. Ozawa, C. Miyamoto, Y. Maehata, A. Suzuki, T. Maeda, and Y. Baba, "Acidic extracellular microenvironment and cancer", Cancer Cell International, vol. 13, pp. 89, 2013. http://dx.doi.org/10.1186/1475-2867-13-89
- G.A. Martínez-Muñoz, and P. Kane, "Vacuolar and Plasma Membrane Proton Pumps Collaborate to Achieve Cytosolic pH Homeostasis in Yeast", Journal of Biological Chemistry, vol. 283, pp. 20309-20319, 2008. http://dx.doi.org/10.1074/jbc.M710470200
- M.S. Cyert, and C.C. Philpott, "Regulation of Cation Balance inSaccharomyces cerevisiae", Genetics, vol. 193, pp. 677-713, 2013. http://dx.doi.org/10.1534/genetics.112.147207
- T. Nishi, and M. Forgac, "The vacuolar (H+)-ATPases — nature's most versatile proton pumps", Nature Reviews Molecular Cell Biology, vol. 3, pp. 94-103, 2002. http://dx.doi.org/10.1038/nrm729
- P.M. Kane, "Proton Transport and pH Control in Fungi", Advances in Experimental Medicine and Biology, pp. 33-68, 2016. http://dx.doi.org/10.1007/978-3-319-25304-6_3
- P.M. Kane, "Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo.", The Journal of biological chemistry, 1995. http://www.ncbi.nlm.nih.gov/pubmed/7622524
- R. Dechant, M. Binda, S.S. Lee, S. Pelet, J. Winderickx, and M. Peter, "Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase", The EMBO Journal, vol. 29, pp. 2515-2526, 2010. http://dx.doi.org/10.1038/emboj.2010.138
- T.T. Diakov, and P.M. Kane, "Regulation of Vacuolar Proton-translocating ATPase Activity and Assembly by Extracellular pH", Journal of Biological Chemistry, vol. 285, pp. 23771-23778, 2010. http://dx.doi.org/10.1074/jbc.M110.110122
- S. Padilla-López, and D.A. Pearce, "Saccharomyces cerevisiae Lacking Btn1p Modulate Vacuolar ATPase Activity to Regulate pH Imbalance in the Vacuole", Journal of Biological Chemistry, vol. 281, pp. 10273-10280, 2006. http://dx.doi.org/10.1074/jbc.M510625200
- R. Dechant, and M. Peter, "The N-terminal domain of the V-ATPase subunit 'a' is regulated by pH in vitro and in vivo", Channels, vol. 5, pp. 4-8, 2011. http://dx.doi.org/10.4161/chan.5.1.13846
- V. Marshansky, "The V-ATPase a2-subunit as a putative endosomal pH-sensor", Biochemical Society Transactions, vol. 35, pp. 1092-1099, 2007. http://dx.doi.org/10.1042/BST0351092
- S. Kawasaki-Nishi, K. Bowers, T. Nishi, M. Forgac, and T.H. Stevens, "The Amino-terminal Domain of the Vacuolar Proton-translocating ATPase a Subunit Controls Targeting and in Vivo Dissociation, and the Carboxyl-terminal Domain Affects Coupling of Proton Transport and ATP Hydrolysis", Journal of Biological Chemistry, vol. 276, pp. 47411-47420, 2001. http://dx.doi.org/10.1074/jbc.M108310200
- L.M. Compton, O.C. Ikonomov, D. Sbrissa, P. Garg, and A. Shisheva, "Active vacuolar H+ ATPase and functional cycle of Rab5 are required for the vacuolation defect triggered by PtdIns(3,5)P2 loss under PIKfyve or Vps34 deficiency", American Journal of Physiology-Cell Physiology, vol. 311, pp. C366-C377, 2016. http://dx.doi.org/10.1152/ajpcell.00104.2016
- S.C. Li, T.T. Diakov, T. Xu, M. Tarsio, W. Zhu, S. Couoh-Cardel, L.S. Weisman, and P.M. Kane, "The signaling lipid PI(3,5)P2stabilizes V1–Vosector interactions and activates the V-ATPase", Molecular Biology of the Cell, vol. 25, pp. 1251-1262, 2014. http://dx.doi.org/10.1091/mbc.E13-10-0563
- S. Banerjee, and P.M. Kane, "Direct interaction of the Golgi V-ATPase a-subunit isoform with PI(4)P drives localization of Golgi V-ATPases in yeast", Molecular Biology of the Cell, vol. 28, pp. 2518-2530, 2017. http://dx.doi.org/10.1091/mbc.E17-05-0316
- S.R. Davis-Kaplan, M.A. Compton, A.R. Flannery, D.M. Ward, J. Kaplan, T.H. Stevens, and L.A. Graham, "PKR1Encodes an Assembly Factor for the Yeast V-Type ATPase", Journal of Biological Chemistry, vol. 281, pp. 32025-32035, 2006. http://dx.doi.org/10.1074/jbc.M606451200
- L.A. Graham, A.R. Flannery, and T.H. Stevens, "Structure and assembly of the yeast V-ATPase.", Journal of bioenergetics and biomembranes, 2003. http://www.ncbi.nlm.nih.gov/pubmed/14635776
- P. Malkus, L.A. Graham, T.H. Stevens, and R. Schekman, "Role of Vma21p in Assembly and Transport of the Yeast Vacuolar ATPase", Molecular Biology of the Cell, vol. 15, pp. 5075-5091, 2004. http://dx.doi.org/10.1091/mbc.E04-06-0514
- M. Ryan, L.A. Graham, and T.H. Stevens, "Voa1p Functions in V-ATPase Assembly in the Yeast Endoplasmic Reticulum", Molecular Biology of the Cell, vol. 19, pp. 5131-5142, 2008. http://dx.doi.org/10.1091/mbc.E08-06-0629
- C.L. Brett, L. Kallay, Z. Hua, R. Green, A. Chyou, Y. Zhang, T.R. Graham, M. Donowitz, and R. Rao, "Genome-Wide Analysis Reveals the Vacuolar pH-Stat of Saccharomyces cerevisiae", PLoS ONE, vol. 6, pp. e17619, 2011. http://dx.doi.org/10.1371/journal.pone.0017619
- R. Orij, M.L. Urbanus, F.J. Vizeacoumar, G. Giaever, C. Boone, C. Nislow, S. Brul, and G.J. Smits, "Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHc in Saccharomyces cerevisiae", Genome Biology, vol. 13, pp. R80, 2012. http://dx.doi.org/10.1186/gb-2012-13-9-r80
- R. Serrano, M.C. Kielland-Brandt, and G.R. Fink, "Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases", Nature, vol. 319, pp. 689-693, 1986. http://dx.doi.org/10.1038/319689a0
- F. Portillo, "Regulation of plasma membrane H(+)-ATPase in fungi and plants.", Biochimica et biophysica acta, 2000. http://www.ncbi.nlm.nih.gov/pubmed/10692636
- P. Eraso, M.J. Mazón, and F. Portillo, "Yeast protein kinase Ptk2 localizes at the plasma membrane and phosphorylates in vitro the C-terminal peptide of the H+-ATPase", Biochimica et Biophysica Acta (BBA) - Biomembranes, vol. 1758, pp. 164-170, 2006. http://dx.doi.org/10.1016/j.bbamem.2006.01.010
- E. Estrada, P. Agostinis, J.R. Vandenheede, J. Goris, W. Merlevede, J. François, A. Goffeau, and M. Ghislain, "Phosphorylation of yeast plasma membrane H+-ATPase by casein kinase I.", The Journal of biological chemistry, 1996. http://www.ncbi.nlm.nih.gov/pubmed/8943257
- M.J. Mazón, P. Eraso, and F. Portillo, "Specific phosphoantibodies reveal two phosphorylation sites in yeast Pma1 in response to glucose", FEMS Yeast Research, vol. 15, pp. fov030, 2015. http://dx.doi.org/10.1093/femsyr/fov030
- L. Bouillet, A. Cardoso, E. Perovano, R. Pereira, E. Ribeiro, M. Trópia, L. Fietto, R. Tisi, E. Martegani, I. Castro, and R. Brandão, "The involvement of calcium carriers and of the vacuole in the glucose-induced calcium signaling and activation of the plasma membrane H+-ATPase in Saccharomyces cerevisiae cells", Cell Calcium, vol. 51, pp. 72-81, 2012. http://dx.doi.org/10.1016/j.ceca.2011.10.008
- L. Yenush, S. Merchan, J. Holmes, and R. Serrano, "pH-Responsive, Posttranslational Regulation of the Trk1 Potassium Transporter by the Type 1-Related Ppz1 Phosphatase", Molecular and Cellular Biology, vol. 25, pp. 8683-8692, 2005. http://dx.doi.org/10.1128/MCB.25.19.8683-8692.2005
- C. Huang, and A. Chang, "pH-dependent Cargo Sorting from the Golgi", Journal of Biological Chemistry, vol. 286, pp. 10058-10065, 2011. http://dx.doi.org/10.1074/jbc.M110.197889
- A.R. Fernandes, and I. Sá‐Correia, "Transcription patterns of PMA1 and PMA2 genes and activity of plasma membrane H+‐ATPase in Saccharomyces cerevisiae during diauxic growth and stationary phase", Yeast, vol. 20, pp. 207-219, 2003. http://dx.doi.org/10.1002/yea.957
- A. Schlesser, S. Ulaszewski, M. Ghislain, and A. Goffeau, "A second transport ATPase gene in Saccharomyces cerevisiae.", The Journal of biological chemistry, 1988. http://www.ncbi.nlm.nih.gov/pubmed/2904437
- E. Eskes, M. Deprez, T. Wilms, and J. Winderickx, "pH homeostasis in yeast; the phosphate perspective", Current Genetics, vol. 64, pp. 155-161, 2017. http://dx.doi.org/10.1007/s00294-017-0743-2
- C. Gournas, M. Prévost, E. Krammer, and B. André, "Function and Regulation of Fungal Amino Acid Transporters: Insights from Predicted Structure", Advances in Experimental Medicine and Biology, pp. 69-106, 2016. http://dx.doi.org/10.1007/978-3-319-25304-6_4
- R. Russnak, D. Konczal, and S.L. McIntire, "A Family of Yeast Proteins Mediating Bidirectional Vacuolar Amino Acid Transport", Journal of Biological Chemistry, vol. 276, pp. 23849-23857, 2001. http://dx.doi.org/10.1074/jbc.M008028200
- M. Conrad, J. Schothorst, H.N. Kankipati, G. Van Zeebroeck, M. Rubio-Texeira, and J.M. Thevelein, "Nutrient sensing and signaling in the yeastSaccharomyces cerevisiae", FEMS Microbiology Reviews, vol. 38, pp. 254-299, 2014. http://dx.doi.org/10.1111/1574-6976.12065
- B. Sampaio-Marques, . , W. Burhans, P. Ludovico, . , and . , "Longevity pathways and maintenance of the proteome: the role of autophagy and mitophagy during yeast ageing", Microbial Cell, vol. 1, pp. 118-127, 2014. http://dx.doi.org/10.15698/mic2014.04.136
- B. Smets, R. Ghillebert, P. De Snijder, M. Binda, E. Swinnen, C. De Virgilio, and J. Winderickx, "Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae", Current Genetics, vol. 56, pp. 1-32, 2010. http://dx.doi.org/10.1007/s00294-009-0287-1
- H. Weidberg, F. Moretto, G. Spedale, A. Amon, and F.J. van Werven, "Nutrient Control of Yeast Gametogenesis Is Mediated by TORC1, PKA and Energy Availability", PLOS Genetics, vol. 12, pp. e1006075, 2016. http://dx.doi.org/10.1371/journal.pgen.1006075
- F. Rolland, J.H. De Winde, K. Lemaire, E. Boles, J.M. Thevelein, and J. Winderickx, "Glucose‐induced cAMP signalling in yeast requires both a G‐protein coupled receptor system for extracellular glucose detection and a separable hexose kinase‐dependent sensing process", Molecular Microbiology, vol. 38, pp. 348-358, 2000. http://dx.doi.org/10.1046/j.1365-2958.2000.02125.x
- D. Stracka, S. Jozefczuk, F. Rudroff, U. Sauer, and M.N. Hall, "Nitrogen Source Activates TOR (Target of Rapamycin) Complex 1 via Glutamine and Independently of Gtr/Rag Proteins", Journal of Biological Chemistry, vol. 289, pp. 25010-25020, 2014. http://dx.doi.org/10.1074/jbc.M114.574335
- M. Binda, M. Péli-Gulli, G. Bonfils, N. Panchaud, J. Urban, T.W. Sturgill, R. Loewith, and C. De Virgilio, "The Vam6 GEF Controls TORC1 by Activating the EGO Complex", Molecular Cell, vol. 35, pp. 563-573, 2009. http://dx.doi.org/10.1016/j.molcel.2009.06.033
- K. Powis, and C. De Virgilio, "Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling", Cell Discovery, vol. 2, 2016. http://dx.doi.org/10.1038/celldisc.2015.49
- R. Zoncu, L. Bar-Peled, A. Efeyan, S. Wang, Y. Sancak, and D.M. Sabatini, "mTORC1 Senses Lysosomal Amino Acids Through an Inside-Out Mechanism That Requires the Vacuolar H + -ATPase", Science, vol. 334, pp. 678-683, 2011. http://dx.doi.org/10.1126/science.1207056
- R. Nicastro, A. Sardu, N. Panchaud, and C. De Virgilio, "The Architecture of the Rag GTPase Signaling Network", Biomolecules, vol. 7, pp. 48, 2017. http://dx.doi.org/10.3390/biom7030048
- J. Thomas, Y. Zhang, Y. Wei, J. Cho, L. Morris, H. Wang, and X. Zheng, "Rab1A Is an mTORC1 Activator and a Colorectal Oncogene", Cancer Cell, vol. 26, pp. 754-769, 2014. http://dx.doi.org/10.1016/j.ccell.2014.09.008
- A. Kim, and K.W. Cunningham, "A LAPF/phafin1-like protein regulates TORC1 and lysosomal membrane permeabilization in response to endoplasmic reticulum membrane stress", Molecular Biology of the Cell, vol. 26, pp. 4631-4645, 2015. http://dx.doi.org/10.1091/mbc.E15-08-0581
- M. Tanigawa, and T. Maeda, "An In Vitro TORC1 Kinase Assay That Recapitulates the Gtr-Independent Glutamine-Responsive TORC1 Activation Mechanism on Yeast Vacuoles", Molecular and Cellular Biology, vol. 37, 2017. http://dx.doi.org/10.1128/MCB.00075-17
- J. Urban, A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, J.R. Broach, C. De Virgilio, M.N. Hall, and R. Loewith, "Sch9 Is a Major Target of TORC1 in Saccharomyces cerevisiae", Molecular Cell, vol. 26, pp. 663-674, 2007. http://dx.doi.org/10.1016/j.molcel.2007.04.020
- E. Swinnen, R. Ghillebert, T. Wilms, and J. Winderickx, "Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 14, pp. 17-32, 2013. http://dx.doi.org/10.1111/1567-1364.12097
- F.M. Roelants, "Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9", Microbiology, vol. 150, pp. 3289-3304, 2004. http://dx.doi.org/10.1099/mic.0.27286-0
- K. Voordeckers, M. Kimpe, S. Haesendonckx, W. Louwet, M. Versele, and J.M. Thevelein, "Yeast 3-Phosphoinositide-dependent Protein Kinase-1 (PDK1) Orthologs Pkh1–3 Differentially Regulate Phosphorylation of Protein Kinase A (PKA) and the Protein Kinase B (PKB)/S6K Ortholog Sch9", Journal of Biological Chemistry, vol. 286, pp. 22017-22027, 2011. http://dx.doi.org/10.1074/jbc.M110.200071
- M. Crauwels, M.C.V. Donaton, M.B. Pernambuco, J. Winderickx, J.H. de Winde, and J.M. Thevelein, "The Sch9 protein kinase in the yeast Saccharomyces cerevisiae controls cAPK activity and is required for nitrogen activation of the fermentable-growth-medium-induced (FGM) pathway", Microbiology, vol. 143, pp. 2627-2637, 1997. http://dx.doi.org/10.1099/00221287-143-8-2627
- J. Lu, Y. Lin, J. Sheu, J. Wu, F. Lee, Y. Chen, M. Lin, F. Chiang, T. Tai, S. Berger, Y. Zhao, K. Tsai, H. Zhu, L. Chuang, and J. Boeke, "Acetylation of Yeast AMPK Controls Intrinsic Aging Independently of Caloric Restriction", Cell, vol. 146, pp. 969-979, 2011. http://dx.doi.org/10.1016/j.cell.2011.07.044
- J. Roosen, K. Engelen, K. Marchal, J. Mathys, G. Griffioen, E. Cameroni, J.M. Thevelein, C. De Virgilio, B. De Moor, and J. Winderickx, "PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability", Molecular Microbiology, vol. 55, pp. 862-880, 2004. http://dx.doi.org/10.1111/j.1365-2958.2004.04429.x
- B. Smets, P. De Snijder, K. Engelen, E. Joossens, R. Ghillebert, K. Thevissen, K. Marchal, and J. Winderickx, "Genome-wide expression analysis reveals TORC1-dependent and -independent functions of Sch9", FEMS Yeast Research, vol. 8, pp. 1276-1288, 2008. http://dx.doi.org/10.1111/j.1567-1364.2008.00432.x
- T. Yorimitsu, S. Zaman, J.R. Broach, and D.J. Klionsky, "Protein Kinase A and Sch9 Cooperatively Regulate Induction of Autophagy inSaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 18, pp. 4180-4189, 2007. http://dx.doi.org/10.1091/mbc.E07-05-0485
- I. Pedruzzi, F. Dubouloz, E. Cameroni, V. Wanke, J. Roosen, J. Winderickx, and C. De Virgilio, "TOR and PKA Signaling Pathways Converge on the Protein Kinase Rim15 to Control Entry into G0", Molecular Cell, vol. 12, pp. 1607-1613, 2003. http://dx.doi.org/10.1016/s1097-2765(03)00485-4
- E. Swinnen, V. Wanke, J. Roosen, B. Smets, F. Dubouloz, I. Pedruzzi, E. Cameroni, C. De Virgilio, and J. Winderickx, "Rim15 and the crossroads of nutrient signalling pathways in Saccharomyces cerevisiae", Cell Division, vol. 1, 2006. http://dx.doi.org/10.1186/1747-1028-1-3
- M. Casal, H. Cardoso, and C. Leao, "Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae", Microbiology, vol. 142, pp. 1385-1390, 1996. http://dx.doi.org/10.1099/13500872-142-6-1385
- D. Bracey, C. Holyoak, and P. Coote, "Comparison of the inhibitory effect of sorbic acid and amphotericin B on Saccharomyces cerevisiae: is growth inhibition dependent on reduced intracellular pH?", Journal of Applied Microbiology, vol. 85, pp. 1056-1066, 1998. http://dx.doi.org/10.1111/j.1365-2672.1998.tb05271.x
- M. Mollapour, A. Shepherd, and P.W. Piper, "Novel stress responses facilitate Saccharomyces cerevisiae growth in the presence of the monocarboxylate preservatives", Yeast, vol. 25, pp. 169-177, 2008. http://dx.doi.org/10.1002/yea.1576
- R. Orij, S. Brul, and G.J. Smits, "Intracellular pH is a tightly controlled signal in yeast", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1810, pp. 933-944, 2011. http://dx.doi.org/10.1016/j.bbagen.2011.03.011
- C. Chan, D. Dominguez, and K.J. Parra, "Regulation of Vacuolar H+-ATPase (V-ATPase) Reassembly by Glycolysis Flow in 6-Phosphofructo-1-kinase (PFK-1)-deficient Yeast Cells", Journal of Biological Chemistry, vol. 291, pp. 15820-15829, 2016. http://dx.doi.org/10.1074/jbc.M116.717488
- P.M. Kane, "The Where, When, and How of Organelle Acidification by the Yeast Vacuolar H + -ATPase", Microbiology and Molecular Biology Reviews, vol. 70, pp. 177-191, 2006. http://dx.doi.org/10.1128/mmbr.70.1.177-191.2006
- K.J. Parra, and P.M. Kane, "Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect.", Molecular and cellular biology, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9819393
- T. Wilms, E. Swinnen, E. Eskes, L. Dolz-Edo, A. Uwineza, R. Van Essche, J. Rosseels, P. Zabrocki, E. Cameroni, V. Franssens, C. De Virgilio, G.J. Smits, and J. Winderickx, "The yeast protein kinase Sch9 adjusts V-ATPase assembly/disassembly to control pH homeostasis and longevity in response to glucose availability", PLOS Genetics, vol. 13, pp. e1006835, 2017. http://dx.doi.org/10.1371/journal.pgen.1006835
- R. Orij, J. Postmus, A. Ter Beek, S. Brul, and G.J. Smits, "In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth", Microbiology, vol. 155, pp. 268-278, 2009. http://dx.doi.org/10.1099/mic.0.022038-0
- S. Ramos, M. Balbin, M. Raposo, E. Valle, and L.A. Pardo, "The Mechanism of Intracellular Acidification Induced by Glucose in Saccharomyces cerevisiae", Microbiology, vol. 135, pp. 2413-2422, 1989. http://dx.doi.org/10.1099/00221287-135-9-2413
- R. Dechant, S. Saad, A. Ibáñez, and M. Peter, "Cytosolic pH Regulates Cell Growth through Distinct GTPases, Arf1 and Gtr1, to Promote Ras/PKA and TORC1 Activity", Molecular Cell, vol. 55, pp. 409-421, 2014. http://dx.doi.org/10.1016/j.molcel.2014.06.002
- S. Bond, and M. Forgac, "The Ras/cAMP/Protein Kinase A Pathway Regulates Glucose-dependent Assembly of the Vacuolar (H+)-ATPase in Yeast", Journal of Biological Chemistry, vol. 283, pp. 36513-36521, 2008. http://dx.doi.org/10.1074/jbc.M805232200
- M. Voss, O. Vitavska, B. Walz, H. Wieczorek, and O. Baumann, "Stimulus-induced Phosphorylation of Vacuolar H+-ATPase by Protein Kinase A", Journal of Biological Chemistry, vol. 282, pp. 33735-33742, 2007. http://dx.doi.org/10.1074/jbc.M703368200
- R. Alzamora, R.F. Thali, F. Gong, C. Smolak, H. Li, C.J. Baty, C.A. Bertrand, Y. Auchli, R.A. Brunisholz, D. Neumann, K.R. Hallows, and N.M. Pastor-Soler, "PKA Regulates Vacuolar H+-ATPase Localization and Activity via Direct Phosphorylation of the A Subunit in Kidney Cells", Journal of Biological Chemistry, vol. 285, pp. 24676-24685, 2010. http://dx.doi.org/10.1074/jbc.M110.106278
- T. Williamson, D. Adiamah, J. Schwartz, and L. Stateva, "Exploring the genetic control of glycolytic oscillations in Saccharomyces Cerevisiae", BMC Systems Biology, vol. 6, 2012. http://dx.doi.org/10.1186/1752-0509-6-108
- C. Chan, and K.J. Parra, "Yeast Phosphofructokinase-1 Subunit Pfk2p Is Necessary for pH Homeostasis and Glucose-dependent Vacuolar ATPase Reassembly", Journal of Biological Chemistry, vol. 289, pp. 19448-19457, 2014. http://dx.doi.org/10.1074/jbc.M114.569855
- M. Lu, D. Ammar, H. Ives, F. Albrecht, and S.L. Gluck, "Physical Interaction between Aldolase and Vacuolar H+-ATPase Is Essential for the Assembly and Activity of the Proton Pump", Journal of Biological Chemistry, vol. 282, pp. 24495-24503, 2007. http://dx.doi.org/10.1074/jbc.M702598200
- M. Lu, L.S. Holliday, L. Zhang, W.A. Dunn, and S.L. Gluck, "Interaction between Aldolase and Vacuolar H+-ATPase", Journal of Biological Chemistry, vol. 276, pp. 30407-30413, 2001. http://dx.doi.org/10.1074/jbc.M008768200
- H. Dihazi, R. Kessler, and K. Eschrich, "Glucose-Induced Stimulation of the Ras-cAMP Pathway in Yeast Leads to Multiple Phosphorylations and Activation of 6-Phosphofructo-2-kinase", Biochemistry, vol. 42, pp. 6275-6282, 2003. http://dx.doi.org/10.1021/bi034167r
- J.H. Seol, A. Shevchenko, A. Shevchenko, and R.J. Deshaies, "Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly", Nature Cell Biology, vol. 3, pp. 384-391, 2001. http://dx.doi.org/10.1038/35070067
- A.M. Smardon, M. Tarsio, and P.M. Kane, "The RAVE Complex Is Essential for Stable Assembly of the Yeast V-ATPase", Journal of Biological Chemistry, vol. 277, pp. 13831-13839, 2002. http://dx.doi.org/10.1074/jbc.M200682200
- F. Portillo, and M.J. Mazón, "The Saccharomyces cerevisiae start mutant carrying the cdc25 mutation is defective in activation of plasma membrane ATPase by glucose", Journal of Bacteriology, vol. 168, pp. 1254-1257, 1986. http://dx.doi.org/10.1128/jb.168.3.1254-1257.1986
- S. Ulaszewski, F. Hilger, and A. Goffeau, "Cyclic AMP controls the plasma membrane H+‐ATPase activity from Saccharomyces cerevisiae", FEBS Letters, vol. 245, pp. 131-136, 1989. http://dx.doi.org/10.1016/0014-5793(89)80206-6
- J. Becher dos Passos, M. Vanhalewyn, R. Lopes Brandão, I.M. Castro, J.R. Nicoli, and J.M. Thevelein, "Glucose-induced activation of plasma membrane H+-ATPase in mutants of the yeast Saccharomyces cerevisiae affected in cAMP metabolism, cAMP-dependent protein phosphorylation and the initiation of glycolysis", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1136, pp. 57-67, 1992. http://dx.doi.org/10.1016/0167-4889(92)90085-P
- M.J. MAZON, M.M. BEHRENS, F. PORTILLO, and R. PInoN, "cAMP- and RAS-independent Nutritional Regulation of Plasma-membrane H+-ATPase Activity in Saccharomyces cerevisiae", Microbiology, vol. 135, pp. 1453-1460, 1989. http://dx.doi.org/10.1099/00221287-135-6-1453
-
S. Mahmoud, M.D. Planes, M. Cabedo, C. Trujillo, A. Rienzo, M. Caballero‐Molada, S.C. Sharma, C. Montesinos, J.M. Mulet, and R. Serrano, "
TOR complex 1 regulates the yeast plasma membrane proton pump andpH and potassium homeostasis", FEBS Letters, vol. 591, pp. 1993-2002, 2017. http://dx.doi.org/10.1002/1873-3468.12673 - C.J. Bonangelino, N.L. Catlett, and L.S. Weisman, "Vac7p, a Novel Vacuolar Protein, Is Required for Normal Vacuole Inheritance and Morphology", Molecular and Cellular Biology, vol. 17, pp. 6847-6858, 1997. http://dx.doi.org/10.1128/MCB.17.12.6847
- C.J. Bonangelino, J.J. Nau, J.E. Duex, M. Brinkman, A.E. Wurmser, J.D. Gary, S.D. Emr, and L.S. Weisman, "Osmotic stress–induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p", The Journal of Cell Biology, vol. 156, pp. 1015-1028, 2002. http://dx.doi.org/10.1083/jcb.200201002
- S.K. Dove, R.K. McEwen, A. Mayes, D.C. Hughes, J.D. Beggs, and R.H. Michell, "Vac14 Controls PtdIns(3,5) P 2 Synthesis and Fab1-Dependent Protein Trafficking to the Multivesicular Body", Current Biology, vol. 12, pp. 885-893, 2002. http://dx.doi.org/10.1016/S0960-9822(02)00891-6
- D. Bridges, K. Fisher, S.N. Zolov, T. Xiong, K. Inoki, L.S. Weisman, and A.R. Saltiel, "Rab5 Proteins Regulate Activation and Localization of Target of Rapamycin Complex 1", Journal of Biological Chemistry, vol. 287, pp. 20913-20921, 2012. http://dx.doi.org/10.1074/jbc.M111.334060
- N. Jin, M. Lang, and L. Weisman, "Phosphatidylinositol 3,5-bisphosphate: regulation of cellular events in space and time", Biochemical Society Transactions, vol. 44, pp. 177-184, 2016. http://dx.doi.org/10.1042/BST20150174
- Y. Jin, and L.S. Weisman, "The vacuole/lysosome is required for cell-cycle progression", eLife, vol. 4, 2015. http://dx.doi.org/10.7554/eLife.08160
- S.A. Rudge, D.M. Anderson, and S.D. Emr, "Vacuole Size Control: Regulation of PtdIns(3,5)P2Levels by the Vacuole-associated Vac14-Fig4 Complex, a PtdIns(3,5)P2-specific Phosphatase", Molecular Biology of the Cell, vol. 15, pp. 24-36, 2004. http://dx.doi.org/10.1091/mbc.E03-05-0297
- B. Stauffer, and T. Powers, "Target of rapamycin signaling mediates vacuolar fission caused by endoplasmic reticulum stress inSaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 26, pp. 4618-4630, 2015. http://dx.doi.org/10.1091/mbc.E15-06-0344
- B.P. Young, J.J.H. Shin, R. Orij, J.T. Chao, S.C. Li, X.L. Guan, A. Khong, E. Jan, M.R. Wenk, W.A. Prinz, G.J. Smits, and C.J.R. Loewen, "Phosphatidic Acid Is a pH Biosensor That Links Membrane Biogenesis to Metabolism", Science, vol. 329, pp. 1085-1088, 2010. http://dx.doi.org/10.1126/science.1191026
- C. Purwin, K. Nicolay, W.A. Scheffers, and H. Holzer, "Mechanism of control of adenylate cyclase activity in yeast by fermentable sugars and carbonyl cyanide m-chlorophenylhydrazone.", The Journal of biological chemistry, 1986. http://www.ncbi.nlm.nih.gov/pubmed/3522579
- J.M. Thevelein, "Fermentable sugars and intracellular acidification as specific activators of the RAS‐adenylate cyclase signalling pathway in yeast: the relationship to nutrient‐induced cell cycle control", Molecular Microbiology, vol. 5, pp. 1301-1307, 1991. http://dx.doi.org/10.1111/j.1365-2958.1991.tb00776.x
- J.M. Trevillyan, and M.L. Pall, "Control of cyclic adenosine 3',5'-monophosphate levels by depolarizing agents in fungi.", Journal of bacteriology, 1979. http://www.ncbi.nlm.nih.gov/pubmed/220213
- S. Colombo, "Involvement of distinct G-proteins, Gpa2 and Ras, in glucose-and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae", The EMBO Journal, vol. 17, pp. 3326-3341, 1998. http://dx.doi.org/10.1093/emboj/17.12.3326
- F. Rolland, V. Wanke, L. Cauwenberg, P. Ma, E. Boles, M. Vanoni, J.H. Winde, J.M. Thevelein, and J. Winderickx, "The role of hexose transport and phosphorylation in cAMP signalling in the yeastSaccharomyces cerevisiae", FEMS Yeast Research, vol. 1, pp. 33-45, 2001. http://dx.doi.org/10.1111/j.1567-1364.2001.tb00011.x
- V. Longo, G. Shadel, M. Kaeberlein, and B. Kennedy, "Replicative and Chronological Aging in Saccharomyces cerevisiae", Cell Metabolism, vol. 16, pp. 18-31, 2012. http://dx.doi.org/10.1016/j.cmet.2012.06.002
- A. Denoth Lippuner, T. Julou, and Y. Barral, "Budding yeast as a model organism to study the effects of age", FEMS Microbiology Reviews, vol. 38, pp. 300-325, 2014. http://dx.doi.org/10.1111/1574-6976.12060
- P. Dakik, and V.I. Titorenko, "Communications between Mitochondria, the Nucleus, Vacuoles, Peroxisomes, the Endoplasmic Reticulum, the Plasma Membrane, Lipid Droplets, and the Cytosol during Yeast Chronological Aging", Frontiers in Genetics, vol. 7, 2016. http://dx.doi.org/10.3389/fgene.2016.00177
- N. Zhang, and L. Cao, "Starvation signals in yeast are integrated to coordinate metabolic reprogramming and stress response to ensure longevity", Current Genetics, vol. 63, pp. 839-843, 2017. http://dx.doi.org/10.1007/s00294-017-0697-4
- F. Tsang, and S. Lin, "Less is more: Nutrient limitation induces cross-talk of nutrient sensing pathways with NAD+ homeostasis and contributes to longevity", Frontiers in Biology, vol. 10, pp. 333-357, 2015. http://dx.doi.org/10.1007/s11515-015-1367-x
- A. Arlia-Ciommo, A. Piano, A. Leonov, V. Svistkova, and V.I. Titorenko, "Quasi-programmed aging of budding yeast: a trade-off between programmed processes of cell proliferation, differentiation, stress response, survival and death defines yeast lifespan", Cell Cycle, vol. 13, pp. 3336-3349, 2014. http://dx.doi.org/10.4161/15384101.2014.965063
- F. Madeo, A. Zimmermann, M.C. Maiuri, and G. Kroemer, "Essential role for autophagy in life span extension", Journal of Clinical Investigation, vol. 125, pp. 85-93, 2015. http://dx.doi.org/10.1172/JCI73946
- A. Bitto, A.M. Wang, C.F. Bennett, and M. Kaeberlein, "Biochemical Genetic Pathways that Modulate Aging in Multiple Species: Figure 1.", Cold Spring Harbor Perspectives in Medicine, vol. 5, pp. a025114, 2015. http://dx.doi.org/10.1101/cshperspect.a025114
- P. Ludovico, and W.C. Burhans, "Reactive oxygen species, ageing and the hormesis police", FEMS Yeast Research, vol. 14, pp. 33-39, 2013. http://dx.doi.org/10.1111/1567-1364.12070
- M.B. Wierman, and J.S. Smith, "Yeast sirtuins and the regulation of aging", FEMS Yeast Research, vol. 14, pp. 73-88, 2013. http://dx.doi.org/10.1111/1567-1364.12115
- J.R. Delaney, C. Murakami, A. Chou, D. Carr, J. Schleit, G.L. Sutphin, E.H. An, A.S. Castanza, M. Fletcher, S. Goswami, S. Higgins, M. Holmberg, J. Hui, M. Jelic, K. Jeong, J.R. Kim, S. Klum, E. Liao, M.S. Lin, W. Lo, H. Miller, R. Moller, Z.J. Peng, T. Pollard, P. Pradeep, D. Pruett, D. Rai, V. Ros, A. Schuster, M. Singh, B.L. Spector, H.V. Wende, A.M. Wang, B.M. Wasko, B. Olsen, and M. Kaeberlein, "Dietary restriction and mitochondrial function link replicative and chronological aging in Saccharomyces cerevisiae", Experimental Gerontology, vol. 48, pp. 1006-1013, 2013. http://dx.doi.org/10.1016/j.exger.2012.12.001
- C. Murakami, J.R. Delaney, A. Chou, D. Carr, J. Schleit, G.L. Sutphin, E.H. An, A.S. Castanza, M. Fletcher, S. Goswami, S. Higgins, M. Holmberg, J. Hui, M. Jelic, K. Jeong, J.R. Kim, S. Klum, E. Liao, M.S. Lin, W. Lo, H. Miller, R. Moller, Z.J. Peng, T. Pollard, P. Pradeep, D. Pruett, D. Rai, V. Ros, A. Schuster, M. Singh, B.L. Spector, H. Vander Wende, A.M. Wang, B.M. Wasko, B. Olsen, and M. Kaeberlein, "pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast", Cell Cycle, vol. 11, pp. 3087-3096, 2012. http://dx.doi.org/10.4161/cc.21465
- M.G. Mirisola, and V.D. Longo, "Acetic acid and acidification accelerate chronological and replicative aging in yeast", Cell Cycle, vol. 11, pp. 3532-3533, 2012. http://dx.doi.org/10.4161/cc.22042
- I. Orlandi, R. Ronzulli, N. Casatta, and M. Vai, "Ethanol and Acetate Acting as Carbon/Energy Sources Negatively Affect Yeast Chronological Aging", Oxidative Medicine and Cellular Longevity, vol. 2013, pp. 1-10, 2013. http://dx.doi.org/10.1155/2013/802870
- K.C. Thomas, S.H. Hynes, and W.M. Ingledew, "Influence of Medium Buffering Capacity on Inhibition ofSaccharomyces cerevisiaeGrowth by Acetic and Lactic Acids", Applied and Environmental Microbiology, vol. 68, pp. 1616-1623, 2002. http://dx.doi.org/10.1128/AEM.68.4.1616-1623.2002
- K.A. Henderson, A.L. Hughes, and D.E. Gottschling, "Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast", eLife, vol. 3, 2014. http://dx.doi.org/10.7554/eLife.03504
- A.M. Smardon, and P.M. Kane, "Loss of Vacuolar H+-ATPase Activity in Organelles Signals Ubiquitination and Endocytosis of the Yeast Plasma Membrane Proton pump Pma1p", Journal of Biological Chemistry, vol. 289, pp. 32316-32326, 2014. http://dx.doi.org/10.1074/jbc.M114.574442
- A. Święciło, "Cross-stress resistance in Saccharomyces cerevisiae yeast—new insight into an old phenomenon", Cell Stress and Chaperones, vol. 21, pp. 187-200, 2016. http://dx.doi.org/10.1007/s12192-016-0667-7
- E.J. Calabrese, K.A. Bachmann, A.J. Bailer, P.M. Bolger, J. Borak, L. Cai, N. Cedergreen, M.G. Cherian, C.C. Chiueh, T.W. Clarkson, R.R. Cook, D.M. Diamond, D.J. Doolittle, M.A. Dorato, S.O. Duke, L. Feinendegen, D.E. Gardner, R.W. Hart, K.L. Hastings, A.W. Hayes, G.R. Hoffmann, J.A. Ives, Z. Jaworowski, T.E. Johnson, W.B. Jonas, N.E. Kaminski, J.G. Keller, J.E. Klaunig, T.B. Knudsen, W.J. Kozumbo, T. Lettieri, S. Liu, A. Maisseu, K.I. Maynard, E.J. Masoro, R.O. McClellan, H.M. Mehendale, C. Mothersill, D.B. Newlin, H.N. Nigg, F.W. Oehme, R.F. Phalen, M.A. Philbert, S.I. Rattan, J.E. Riviere, J. Rodricks, R.M. Sapolsky, B.R. Scott, C. Seymour, D.A. Sinclair, J. Smith-Sonneborn, E.T. Snow, L. Spear, D.E. Stevenson, Y. Thomas, M. Tubiana, G.M. Williams, and M.P. Mattson, "Biological stress response terminology: Integrating the concepts of adaptive response and preconditioning stress within a hormetic dose–response framework", Toxicology and Applied Pharmacology, vol. 222, pp. 122-128, 2007. http://dx.doi.org/10.1016/j.taap.2007.02.015
- S. Giannattasio, N. Guaragnella, M. Corte-Real, S. Passarella, and E. Marra, "Acid stress adaptation protects Saccharomyces cerevisiae from acetic acid-induced programmed cell death", Gene, vol. 354, pp. 93-98, 2005. http://dx.doi.org/10.1016/j.gene.2005.03.030
- G.R. Hoffmann, A.V. Moczula, A.M. Laterza, L.K. MacNeil, and J.P. Tartaglione, "Adaptive response to hydrogen peroxide in yeast: Induction, time course, and relationship to dose–response models", Environmental and Molecular Mutagenesis, vol. 54, pp. 384-396, 2013. http://dx.doi.org/10.1002/em.21785
- A. Mesquita, M. Weinberger, A. Silva, B. Sampaio-Marques, B. Almeida, C. Leão, V. Costa, F. Rodrigues, W.C. Burhans, and P. Ludovico, "Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H 2 O 2 and superoxide dismutase activity", Proceedings of the National Academy of Sciences, vol. 107, pp. 15123-15128, 2010. http://dx.doi.org/10.1073/pnas.1004432107
- C. Casado, A. González, M. Platara, A. Ruiz, and J. Ariño, "The role of the protein kinase A pathway in the response to alkaline pH stress in yeast", Biochemical Journal, vol. 438, pp. 523-533, 2011. http://dx.doi.org/10.1042/BJ20110607
- N.P. Mira, J.D. Becker, and I. Sá-Correia, "Genomic Expression Program Involving the Haa1p-Regulon inSaccharomyces cerevisiaeResponse to Acetic Acid", OMICS: A Journal of Integrative Biology, vol. 14, pp. 587-601, 2010. http://dx.doi.org/10.1089/omi.2010.0048
- N.P. Mira, M.C. Teixeira, and I. Sá-Correia, "Adaptive Response and Tolerance to Weak Acids inSaccharomyces cerevisiae: A Genome-Wide View", OMICS: A Journal of Integrative Biology, vol. 14, pp. 525-540, 2010. http://dx.doi.org/10.1089/omi.2010.0072
- P. Ludovico, M.J. Sousa, M.T. Silva, C. Leão, and M. Côrte-Real, "Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid", Microbiology, vol. 147, pp. 2409-2415, 2001. http://dx.doi.org/10.1099/00221287-147-9-2409
- P. Ludovico, F. Rodrigues, A. Almeida, M.T. Silva, A. Barrientos, and M. Côrte-Real, "CytochromecRelease and Mitochondria Involvement in Programmed Cell Death Induced by Acetic Acid inSaccharomyces cerevisiae", Molecular Biology of the Cell, vol. 13, pp. 2598-2606, 2002. http://dx.doi.org/10.1091/mbc.E01-12-0161
- N. Guaragnella, M. Ždralević, L. Antonacci, S. Passarella, E. Marra, and S. Giannattasio, "The role of mitochondria in yeast programmed cell death", Frontiers in Oncology, vol. 2, 2012. http://dx.doi.org/10.3389/fonc.2012.00070
- Y. Pan, "Mitochondria, reactive oxygen species, and chronological aging: A message from yeast", Experimental Gerontology, vol. 46, pp. 847-852, 2011. http://dx.doi.org/10.1016/j.exger.2011.08.007
- A.M. Aerts, P. Zabrocki, G. Govaert, J. Mathys, D. Carmona-Gutierrez, F. Madeo, J. Winderickx, B.P. Cammue, and K. Thevissen, "Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast", FEBS Letters, vol. 583, pp. 113-117, 2008. http://dx.doi.org/10.1016/j.febslet.2008.11.028
- P. Fabrizio, L. Battistella, R. Vardavas, C. Gattazzo, L. Liou, A. Diaspro, J.W. Dossen, E.B. Gralla, and V.D. Longo, "Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae ", The Journal of Cell Biology, vol. 166, pp. 1055-1067, 2004. http://dx.doi.org/10.1083/jcb.200404002
- A.L. Hughes, and D.E. Gottschling, "An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast", Nature, vol. 492, pp. 261-265, 2012. http://dx.doi.org/10.1038/nature11654
- P. Laun, A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K. Fröhlich, and M. Breitenbach, "Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis", Molecular Microbiology, vol. 39, pp. 1166-1173, 2001. http://dx.doi.org/10.1111/j.1365-2958.2001.02317.x
- S. Jazwinski, "Mitochondria to nucleus signaling and the role of ceramide in its integration into the suite of cell quality control processes during aging", Ageing Research Reviews, vol. 23, pp. 67-74, 2015. http://dx.doi.org/10.1016/j.arr.2014.12.007
- A. Traven, J.M. Wong, D. Xu, M. Sopta, and C.J. Ingles, "Interorganellar Communication", Journal of Biological Chemistry, vol. 276, pp. 4020-4027, 2001. http://dx.doi.org/10.1074/jbc.M006807200
- J. Hu, M. Wei, H. Mirzaei, F. Madia, M. Mirisola, C. Amparo, S. Chagoury, B. Kennedy, and V.D. Longo, "Tor‐Sch9 deficiency activates catabolism of the ketone body‐like acetic acid to promote trehalose accumulation and longevity", Aging Cell, vol. 13, pp. 457-467, 2014. http://dx.doi.org/10.1111/acel.12202
- M. Petitjean, M. Teste, J.M. François, and J. Parrou, "Yeast Tolerance to Various Stresses Relies on the Trehalose-6P Synthase (Tps1) Protein, Not on Trehalose", Journal of Biological Chemistry, vol. 290, pp. 16177-16190, 2015. http://dx.doi.org/10.1074/jbc.M115.653899
- M. Petitjean, M. Teste, I. Léger-Silvestre, J.M. François, and J. Parrou, "RETRACTED: A new function for the yeast trehalose-6P synthase (Tps1) protein, as key pro-survival factor during growth, chronological ageing, and apoptotic stress", Mechanisms of Ageing and Development, vol. 161, pp. 234-246, 2017. http://dx.doi.org/10.1016/j.mad.2016.07.011
- P. Kyryakov, A. Beach, V.R. Richard, M.T. Burstein, A. Leonov, S. Levy, and V.I. Titorenko, "Caloric Restriction Extends Yeast Chronological Lifespan by Altering a Pattern of Age-Related Changes in Trehalose Concentration", Frontiers in Physiology, vol. 3, 2012. http://dx.doi.org/10.3389/fphys.2012.00256
- N. Nakamura, A. Matsuura, Y. Wada, and Y. Ohsumi, "Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae.", Journal of biochemistry, 1997. http://www.ncbi.nlm.nih.gov/pubmed/9089409
- C. Ruckenstuhl, C. Netzberger, I. Entfellner, D. Carmona-Gutierrez, T. Kickenweiz, S. Stekovic, C. Gleixner, C. Schmid, L. Klug, A.G. Sorgo, T. Eisenberg, S. Büttner, G. Mariño, R. Koziel, P. Jansen-Dürr, K. Fröhlich, G. Kroemer, and F. Madeo, "Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification", PLoS Genetics, vol. 10, pp. e1004347, 2014. http://dx.doi.org/10.1371/journal.pgen.1004347
- F. Tang, J.W. Watkins, M. Bermudez, R. Gray, A. Gaban, K. Portie, S. Grace, M. Kleve, and G. Craciun, "A lifespan-extending form of autophagy employs the vacuole-vacuole fusion machinery", Autophagy, vol. 4, pp. 874-886, 2008. http://dx.doi.org/10.4161/auto.6556
- T. Eisenberg, H. Knauer, A. Schauer, S. Büttner, C. Ruckenstuhl, D. Carmona-Gutierrez, J. Ring, S. Schroeder, C. Magnes, L. Antonacci, H. Fussi, L. Deszcz, R. Hartl, E. Schraml, A. Criollo, E. Megalou, D. Weiskopf, P. Laun, G. Heeren, M. Breitenbach, B. Grubeck-Loebenstein, E. Herker, B. Fahrenkrog, K. Fröhlich, F. Sinner, N. Tavernarakis, N. Minois, G. Kroemer, and F. Madeo, "Induction of autophagy by spermidine promotes longevity", Nature Cell Biology, vol. 11, pp. 1305-1314, 2009. http://dx.doi.org/10.1038/ncb1975
- S.B. Miller, C. Ho, J. Winkler, M. Khokhrina, A. Neuner, M.Y. Mohamed, D.L. Guilbride, K. Richter, M. Lisby, E. Schiebel, A. Mogk, and B. Bukau, "Compartment‐specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition", The EMBO Journal, vol. 34, pp. 778-797, 2015. http://dx.doi.org/10.15252/embj.201489524
- D. Kaganovich, R. Kopito, and J. Frydman, "Misfolded proteins partition between two distinct quality control compartments", Nature, vol. 454, pp. 1088-1095, 2008. http://dx.doi.org/10.1038/nature07195
- J. Song, Q. Yang, J. Yang, L. Larsson, X. Hao, X. Zhu, S. Malmgren-Hill, M. Cvijovic, J. Fernandez-Rodriguez, J. Grantham, C.M. Gustafsson, B. Liu, and T. Nyström, "Essential Genetic Interactors of SIR2 Required for Spatial Sequestration and Asymmetrical Inheritance of Protein Aggregates", PLoS Genetics, vol. 10, pp. e1004539, 2014. http://dx.doi.org/10.1371/journal.pgen.1004539
- B. Liu, L. Larsson, A. Caballero, X. Hao, D. Öling, J. Grantham, and T. Nyström, "The Polarisome Is Required for Segregation and Retrograde Transport of Protein Aggregates", Cell, vol. 140, pp. 257-267, 2010. http://dx.doi.org/10.1016/j.cell.2009.12.031
- B. Liu, L. Larsson, V. Franssens, X. Hao, S. Hill, V. Andersson, D. Höglund, J. Song, X. Yang, D. Öling, J. Grantham, J. Winderickx, and T. Nyström, "Segregation of Protein Aggregates Involves Actin and the Polarity Machinery", Cell, vol. 147, pp. 959-961, 2011. http://dx.doi.org/10.1016/j.cell.2011.11.018
- Y. Liu, N. Liu, D. Wu, Q. Bi, and S. Meng, "The longevity of tor1Δ, sch9Δ, and ras2Δ mutants depends on actin dynamics in Saccharomyces cerevisiae", Cell & Bioscience, vol. 5, 2015. http://dx.doi.org/10.1186/s13578-015-0008-z
- D. Amberg, J.E. Leadsham, V. Kotiadis, and C.W. Gourlay, "Cellular Ageing and the Actin Cytoskeleton", Aging Research in Yeast, pp. 331-352, 2011. http://dx.doi.org/10.1007/978-94-007-2561-4_15
- S. Büttner, C. Delay, V. Franssens, T. Bammens, D. Ruli, S. Zaunschirm, R.M. de Oliveira, T.F. Outeiro, F. Madeo, L. Buée, M. Galas, and J. Winderickx, "Synphilin-1 Enhances α-Synuclein Aggregation in Yeast and Contributes to Cellular Stress and Cell Death in a Sir2-Dependent Manner", PLoS ONE, vol. 5, pp. e13700, 2010. http://dx.doi.org/10.1371/journal.pone.0013700
- S. Hill, X. Hao, J. Grönvall, S. Spikings-Nordby, P. Widlund, T. Amen, A. Jörhov, R. Josefson, D. Kaganovich, B. Liu, and T. Nyström, "Asymmetric Inheritance of Aggregated Proteins and Age Reset in Yeast Are Regulated by Vac17-Dependent Vacuolar Functions", Cell Reports, vol. 16, pp. 826-838, 2016. http://dx.doi.org/10.1016/j.celrep.2016.06.016
- R. Higuchi-Sanabria, W.M. Pernice, J.D. Vevea, D.M. Alessi Wolken, I.R. Boldogh, and L.A. Pon, "Role of asymmetric cell division in lifespan control inSaccharomyces cerevisiae", FEMS Yeast Research, vol. 14, pp. 1133-1146, 2014. http://dx.doi.org/10.1111/1567-1364.12216
- M. Perić, P.B. Dib, S. Dennerlein, M. Musa, M. Rudan, A. Lovrić, A. Nikolić, A. Šarić, S. Sobočanec, �. Mačak, N. Raimundo, and A. Kriško, "Crosstalk between cellular compartments protects against proteotoxicity and extends lifespan", Scientific Reports, vol. 6, 2016. http://dx.doi.org/10.1038/srep28751
-
M. Perić, A. Lovrić, A. Šarić, M. Musa, P. Bou Dib, M. Rudan, A. Nikolić, S. Sobočanec, A. Mikecin, S. Dennerlein, I. Milošević, K. Vlahoviček, N. Raimundo, and A. Kriško, "
TORC 1‐mediated sensing of chaperone activity alters glucose metabolism and extends lifespan", Aging Cell, vol. 16, pp. 994-1005, 2017. http://dx.doi.org/10.1111/acel.12623 - B. Meusser, C. Hirsch, E. Jarosch, and T. Sommer, "ERAD: the long road to destruction", Nature Cell Biology, vol. 7, pp. 766-772, 2005. http://dx.doi.org/10.1038/ncb0805-766
- E. Taylor, and J. Rutter, "Mitochondrial quality control by the ubiquitin–proteasome system", Biochemical Society Transactions, vol. 39, pp. 1509-1513, 2011. http://dx.doi.org/10.1042/BST0391509
- A. Lang, A.T. John Peter, and B. Kornmann, "ER–mitochondria contact sites in yeast: beyond the myths of ERMES", Current Opinion in Cell Biology, vol. 35, pp. 7-12, 2015. http://dx.doi.org/10.1016/j.ceb.2015.03.002
- Y. Elbaz-Alon, E. Rosenfeld-Gur, V. Shinder, A. Futerman, T. Geiger, and M. Schuldiner, "A Dynamic Interface between Vacuoles and Mitochondria in Yeast", Developmental Cell, vol. 30, pp. 95-102, 2014. http://dx.doi.org/10.1016/j.devcel.2014.06.007
- C. Hönscher, M. Mari, K. Auffarth, M. Bohnert, J. Griffith, W. Geerts, M. van der Laan, M. Cabrera, F. Reggiori, and C. Ungermann, "Cellular Metabolism Regulates Contact Sites between Vacuoles and Mitochondria", Developmental Cell, vol. 30, pp. 86-94, 2014. http://dx.doi.org/10.1016/j.devcel.2014.06.006
- N. Belgareh-Touzé, L. Cavellini, and M.M. Cohen, "Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy", Autophagy, vol. 13, pp. 114-132, 2017. http://dx.doi.org/10.1080/15548627.2016.1252889
- S. Böckler, and B. Westermann, "Mitochondrial ER Contacts Are Crucial for Mitophagy in Yeast", Developmental Cell, vol. 28, pp. 450-458, 2014. http://dx.doi.org/10.1016/j.devcel.2014.01.012
- S. Böckler, and B. Westermann, "ER-mitochondria contacts as sites of mitophagosome formation", Autophagy, vol. 10, pp. 1346-1347, 2014. http://dx.doi.org/10.4161/auto.28981
-
R. Lakhani, K.R. Vogel, A. Till, J. Liu, S.F. Burnett, K.M. Gibson, and S. Subramani, "Defects in
GABA metabolism affect selective autophagy pathways and are alleviated by mTOR inhibition", EMBO Molecular Medicine, vol. 6, pp. 551-566, 2014. http://dx.doi.org/10.1002/emmm.201303356 - V.Y. Nazarko, J.M. Thevelein, and A.A. Sibirny, "G‐protein‐coupled receptor Gpr1 and G‐protein Gpa2 of cAMP‐dependent signaling pathway are involved in glucose‐induced pexophagy in the yeast Saccharomyces cerevisiae", Cell Biology International, vol. 32, pp. 502-504, 2008. http://dx.doi.org/10.1016/j.cellbi.2007.11.001
- K. Natter, P. Leitner, A. Faschinger, H. Wolinski, S. McCraith, S. Fields, and S.D. Kohlwein, "The Spatial Organization of Lipid Synthesis in the Yeast Saccharomyces cerevisiae Derived from Large Scale Green Fluorescent Protein Tagging and High Resolution Microscopy", Molecular & Cellular Proteomics, vol. 4, pp. 662-672, 2005. http://dx.doi.org/10.1074/mcp.M400123-MCP200
- S. Bahmanyar, "Spatial regulation of phospholipid synthesis within the nuclear envelope domain of the endoplasmic reticulum", Nucleus, vol. 6, pp. 102-106, 2015. http://dx.doi.org/10.1080/19491034.2015.1010942
- J. Fernández-Murray, and C.R. McMaster, "Lipid synthesis and membrane contact sites: a crossroads for cellular physiology", Journal of Lipid Research, vol. 57, pp. 1789-1805, 2016. http://dx.doi.org/10.1194/jlr.R070920
- P. Malia, and C. Ungermann, "Vacuole membrane contact sites and domains: emerging hubs to coordinate organelle function with cellular metabolism", Biochemical Society Transactions, vol. 44, pp. 528-533, 2016. http://dx.doi.org/10.1042/BST20150277
- A. Murley, R.D. Sarsam, A. Toulmay, J. Yamada, W.A. Prinz, and J. Nunnari, "Ltc1 is an ER-localized sterol transporter and a component of ER–mitochondria and ER–vacuole contacts", Journal of Cell Biology, vol. 209, pp. 539-548, 2015. http://dx.doi.org/10.1083/jcb.201502033
- N. Shai, M. Schuldiner, and E. Zalckvar, "No peroxisome is an island — Peroxisome contact sites", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1863, pp. 1061-1069, 2016. http://dx.doi.org/10.1016/j.bbamcr.2015.09.016
- S. Tavassoli, J.T. Chao, B.P. Young, R.C. Cox, W.A. Prinz, A.I.P.M. de Kroon, and C.J.R. Loewen, "Plasma membrane—endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis", EMBO reports, vol. 14, pp. 434-440, 2013. http://dx.doi.org/10.1038/embor.2013.36
- M.A. De Matteis, and L.R. Rega, "Endoplasmic reticulum–Golgi complex membrane contact sites", Current Opinion in Cell Biology, vol. 35, pp. 43-50, 2015. http://dx.doi.org/10.1016/j.ceb.2015.04.001
- W.M. Henne, J. Liou, and S.D. Emr, "Molecular mechanisms of inter-organelle ER–PM contact sites", Current Opinion in Cell Biology, vol. 35, pp. 123-130, 2015. http://dx.doi.org/10.1016/j.ceb.2015.05.001
- W. Wolf, A. Kilic, B. Schrul, H. Lorenz, B. Schwappach, and M. Seedorf, "Yeast Ist2 Recruits the Endoplasmic Reticulum to the Plasma Membrane and Creates a Ribosome-Free Membrane Microcompartment", PLoS ONE, vol. 7, pp. e39703, 2012. http://dx.doi.org/10.1371/journal.pone.0039703
- E.B. Yucel, S. Eraslan, and K.O. Ulgen, "The impact of medium acidity on the chronological life span of Saccharomyces cerevisiae – lipids, signaling cascades, mitochondrial and vacuolar functions", The FEBS Journal, vol. 281, pp. 1281-1303, 2014. http://dx.doi.org/10.1111/febs.12705
- D. Mitrofanova, P. Dakik, M. McAuley, Y. Medkour, K. Mohammad, and V.I. Titorenko, "Lipid metabolism and transport define longevity of the yeast Saccharomyces cerevisiae.", Frontiers in bioscience (Landmark edition), 2018. http://www.ncbi.nlm.nih.gov/pubmed/28930594
- V.R. Richard, A. Beach, A. Piano, A. Leonov, R. Feldman, M.T. Burstein, P. Kyryakov, A. Gomez-Perez, A. Arlia-Ciommo, S. Baptista, C. Campbell, D. Goncharov, S. Pannu, D. Patrinos, B. Sadri, V. Svistkova, A. Victor, and V.I. Titorenko, "Mechanism of liponecrosis, a distinct mode of programmed cell death", Cell Cycle, vol. 13, pp. 3707-3726, 2014. http://dx.doi.org/10.4161/15384101.2014.965003
- T. Eisenberg, and S. Büttner, "Lipids and cell death in yeast", FEMS Yeast Research, vol. 14, pp. 179-197, 2014. http://dx.doi.org/10.1111/1567-1364.12105
- K. Halbleib, K. Pesek, R. Covino, H.F. Hofbauer, D. Wunnicke, I. Hänelt, G. Hummer, and R. Ernst, "Activation of the Unfolded Protein Response by Lipid Bilayer Stress", Molecular Cell, vol. 67, pp. 673-684.e8, 2017. http://dx.doi.org/10.1016/j.molcel.2017.06.012
- T. Promlek, Y. Ishiwata-Kimata, M. Shido, M. Sakuramoto, K. Kohno, and Y. Kimata, "Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways", Molecular Biology of the Cell, vol. 22, pp. 3520-3532, 2011. http://dx.doi.org/10.1091/mbc.E11-04-0295
- K. Obara, and A. Kihara, "The Rim101 pathway contributes to ER stress adaptation through sensing the state of plasma membrane", Biochemical Journal, vol. 474, pp. 51-63, 2016. http://dx.doi.org/10.1042/BCJ20160580
- L. Pineau, J. Colas, S. Dupont, L. Beney, P. Fleurat‐Lessard, J. Berjeaud, T. Bergès, and T. Ferreira, "Lipid‐Induced ER Stress: Synergistic Effects of Sterols and Saturated Fatty Acids", Traffic, vol. 10, pp. 673-690, 2009. http://dx.doi.org/10.1111/j.1600-0854.2009.00903.x
- L. Pineau, and T. Ferreira, "Lipid-induced ER stress in yeast and β cells: parallel trails to a common fate", FEMS Yeast Research, vol. 10, pp. 1035-1045, 2010. http://dx.doi.org/10.1111/j.1567-1364.2010.00674.x
- M. Zhang, Q. Yu, C. Liang, B. Zhang, and M. Li, "Lipid homeostasis is involved in plasma membrane and endoplasmic reticulum stress in Pichia pastoris", Biochemical and Biophysical Research Communications, vol. 478, pp. 777-783, 2016. http://dx.doi.org/10.1016/j.bbrc.2016.08.024
- E. Navarro-Tapia, R.K. Nana, A. Querol, and R. Pérez-Torrado, "Ethanol Cellular Defense Induce Unfolded Protein Response in Yeast", Frontiers in Microbiology, vol. 7, 2016. http://dx.doi.org/10.3389/fmicb.2016.00189
- N. Austriaco, O. P., "Endoplasmic reticulum involvement in yeast cell death", Frontiers in Oncology, vol. 2, 2012. http://dx.doi.org/10.3389/fonc.2012.00087
- R.J. Braun, and H. Zischka, "Mechanisms of Cdc48/VCP-mediated cell death — from yeast apoptosis to human disease", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1783, pp. 1418-1435, 2008. http://dx.doi.org/10.1016/j.bbamcr.2008.01.015
- R.D. Moir, D.A. Gross, D.L. Silver, and I.M. Willis, "SCS3 and YFT2 Link Transcription of Phospholipid Biosynthetic Genes to ER Stress and the UPR", PLoS Genetics, vol. 8, pp. e1002890, 2012. http://dx.doi.org/10.1371/journal.pgen.1002890
- S. Epstein, C.L. Kirkpatrick, G.A. Castillon, M. Muñiz, I. Riezman, F.P. David, C.B. Wollheim, and H. Riezman, "Activation of the unfolded protein response pathway causes ceramide accumulation in yeast and INS-1E insulinoma cells", Journal of Lipid Research, vol. 53, pp. 412-420, 2012. http://dx.doi.org/10.1194/jlr.M022186
- S. Jadhav, S. Russo, S. Cottier, R. Schneiter, A. Cowart, and M.L. Greenberg, "Valproate Induces the Unfolded Protein Response by Increasing Ceramide Levels", Journal of Biological Chemistry, vol. 291, pp. 22253-22261, 2016. http://dx.doi.org/10.1074/jbc.M116.752634
- G. Thibault, G. Shui, W. Kim, G. McAlister, N. Ismail, S. Gygi, M. Wenk, and D. Ng, "The Membrane Stress Response Buffers Lethal Effects of Lipid Disequilibrium by Reprogramming the Protein Homeostasis Network", Molecular Cell, vol. 48, pp. 16-27, 2012. http://dx.doi.org/10.1016/j.molcel.2012.08.016
- M. Liu, C. Huang, S.R. Polu, R. Schneiter, and A. Chang, "Regulation of sphingolipid synthesis via Orm1 and Orm2 in yeast", Journal of Cell Science, 2012. http://dx.doi.org/10.1242/jcs.100578
- W. Fei, H. Wang, X. Fu, C. Bielby, and H. Yang, "Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae", Biochemical Journal, vol. 424, pp. 61-67, 2009. http://dx.doi.org/10.1042/BJ20090785
- J. Bischof, M. Salzmann, M.K. Streubel, J. Hasek, F. Geltinger, J. Duschl, N. Bresgen, P. Briza, D. Haskova, R. Lejskova, M. Sopjani, K. Richter, and M. Rinnerthaler, "Clearing the outer mitochondrial membrane from harmful proteins via lipid droplets", Cell Death Discovery, vol. 3, 2017. http://dx.doi.org/10.1038/cddiscovery.2017.16
- T. Shpilka, and Z. Elazar, "Lipid droplets regulate autophagosome biogenesis", Autophagy, vol. 11, pp. 2130-2131, 2015. http://dx.doi.org/10.1080/15548627.2015.1093719
- T. Shpilka, E. Welter, N. Borovsky, N. Amar, M. Mari, F. Reggiori, and Z. Elazar, "Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis", The EMBO Journal, vol. 34, pp. 2117-2131, 2015. http://dx.doi.org/10.15252/embj.201490315
- A.P. Velázquez, T. Tatsuta, R. Ghillebert, I. Drescher, and M. Graef, "Lipid droplet–mediated ER homeostasis regulates autophagy and cell survival during starvation", Journal of Cell Biology, vol. 212, pp. 621-631, 2016. http://dx.doi.org/10.1083/jcb.201508102
- J. Vevea, E. Garcia, R. Chan, B. Zhou, M. Schultz, G. Di Paolo, J. McCaffery, and L. Pon, "Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast", Developmental Cell, vol. 35, pp. 584-599, 2015. http://dx.doi.org/10.1016/j.devcel.2015.11.010
- R.M. Deranieh, Y. Shi, M. Tarsio, Y. Chen, J.M. McCaffery, P.M. Kane, and M.L. Greenberg, "Perturbation of the Vacuolar ATPase", Journal of Biological Chemistry, vol. 290, pp. 27460-27472, 2015. http://dx.doi.org/10.1074/jbc.M115.683706
- A. Hernández, G. Serrano-Bueno, J.R. Perez-Castiñeira, and A. Serrano, "8-Dehydrosterols induce membrane traffic and autophagy defects through V-ATPase dysfunction in Saccharomyces cerevisae", Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, vol. 1853, pp. 2945-2956, 2015. http://dx.doi.org/10.1016/j.bbamcr.2015.09.001
- V. Teixeira, T.C. Medeiros, R. Vilaça, J. Ferreira, P. Moradas-Ferreira, and V. Costa, "Ceramide signaling targets the PP2A-like protein phosphatase Sit4p to impair vacuolar function, vesicular trafficking and autophagy in Isc1p deficient cells", Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, vol. 1861, pp. 21-33, 2016. http://dx.doi.org/10.1016/j.bbalip.2015.10.004
- A.M. Aerts, P. Zabrocki, I.E.J.A. François, D. Carmona-Gutierrez, G. Govaert, C. Mao, B. Smets, F. Madeo, J. Winderickx, B.P.A. Cammue, and K. Thevissen, "Ydc1p ceramidase triggers organelle fragmentation, apoptosis and accelerated ageing in yeast", Cellular and Molecular Life Sciences, vol. 65, pp. 1933-1942, 2008. http://dx.doi.org/10.1007/s00018-008-8129-8
- G.L. Sherr, N. LaMassa, E. Li, G. Phillips, and C. Shen, "Pah1p negatively regulates the expression of V-ATPase genes as well as vacuolar acidification", Biochemical and Biophysical Research Communications, vol. 491, pp. 693-700, 2017. http://dx.doi.org/10.1016/j.bbrc.2017.07.127
- S. Fakas, Y. Qiu, J.L. Dixon, G. Han, K.V. Ruggles, J. Garbarino, S.L. Sturley, and G.M. Carman, "Phosphatidate Phosphatase Activity Plays Key Role in Protection against Fatty Acid-induced Toxicity in Yeast", Journal of Biological Chemistry, vol. 286, pp. 29074-29085, 2011. http://dx.doi.org/10.1074/jbc.M111.258798
- F. Kliewe, J. Kumme, M. Grigat, S. Hintze, and H. Schüller, "Opi1 mediates repression of phospholipid biosynthesis by phosphate limitation in the yeastSaccharomyces cerevisiae", Yeast, vol. 34, pp. 67-81, 2016. http://dx.doi.org/10.1002/yea.3215
ACKNOWLEDGMENTS
We gratefully thank FWO Vlaanderen for a fellowship to EE and for support via the research grants G.0694.13, G.0A63.15 and SBO-S006617N. We also thank KU Leuven for support by granting the C14/17/063 project.
COPYRIGHT
© 2018

pH homeostasis links the nutrient sensing PKA/TORC1/Sch9 ménage-à-trois to stress tolerance and longevity by Deprez et al. is licensed under a Creative Commons Attribution 4.0 International License.