Research Articles:
Microbial Cell, Vol. 2, No. 1, pp. 5 - 13; doi: 10.15698/mic2015.01.182
Prokaryotic ancestry and gene fusion of a dual localized peroxiredoxin in malaria parasites
1 Department of Parasitology, Ruprecht-Karls University, D-69120 Heidelberg, Germany.
2 Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), D-69117 Heidelberg, Germany.
3 Department of Parasitology, Philipps University, D-35043 Marburg, Germany.
Keywords: peroxiredoxin, molecular evolution, horizontal gene transfer, apicomplexa, malaria.
Abbreviations:
BTS - bipartite topogenic signal.
Received originally: 12/12/2014 Accepted: 18/12/2014
Published: 05/01/2015
Correspondence:
Marcel Deponte, Department of Parasitology, Ruprecht-Karls University, Im Neuenheimer Feld 324; D-69120 Heidelberg, Germany marcel.deponte@gmx.de
Conflict of interest statement: The authors declare no conflict of interest.
Please cite this article as: Carine F. Djuika, Jaime Huerta-Cepas, Jude M. Przyborski, Sophia Deil, Cecilia P. Sanchez, Tobias Doerks, Peer Bork, Michael Lanzer, and Marcel Deponte (2015). Prokaryotic ancestry and gene fusion of a dual localized peroxiredoxin in malaria parasites. Microbial Cell 2(1): 5-13.
Abstract
Horizontal gene transfer has emerged as a crucial driving force for the evolution of eukaryotes. This also includes Plasmodium falciparum and related economically and clinically relevant apicomplexan parasites, whose rather small genomes have been shaped not only by natural selection in different host populations but also by horizontal gene transfer following endosymbiosis. However, there is rather little reliable data on horizontal gene transfer between animal hosts or bacteria and apicomplexan parasites. Here we show that apicomplexan homologues of peroxiredoxin 5 (Prx5) have a prokaryotic ancestry and therefore represent a special subclass of Prx5 isoforms in eukaryotes. Using two different immunobiochemical approaches, we found that the P. falciparum Prx5 homologue is dually localized to the parasite plastid and cytosol. This dual localization is reflected by a modular Plasmodium-specific gene architecture consisting of two exons. Despite the plastid localization, our phylogenetic analyses contradict an acquisition by secondary endosymbiosis and support a gene fusion event following a horizontal prokaryote-to-eukaryote gene transfer in early apicomplexans. The results provide unexpected insights into the evolution of apicomplexan parasites as well as the molecular evolution of peroxiredoxins, an important family of ubiquitous, usually highly concentrated thiol-dependent hydroperoxidases that exert functions as detoxifying enzymes, redox sensors and chaperones.
INTRODUCTION
Parasitic lifestyles require dramatic changes of parasite genomes. An important option for such changes is the horizontal transfer of single or multiple genes across regular mating barriers [1][2][3]. Accordingly, horizontal gene transfer following primary and secondary endosymbiosis has significantly altered the genomes of apicomplexan parasites including the human malaria parasite Plasmodium falciparum [4][5][6][7]. In contrast to our knowledge on the acquisition of parasite genes from endosymbionts, only few examples for horizontal gene transfer from animal hosts or bacteria to apicomplexan parasites have been described [7][8][9][10][11][12]. These examples support the importance of horizontal gene transfer for the evolution of apicomplexan parasites with regard to metabolic streamlining [8][9], the coordinated regulation of gene expression and the optimization of cytoadhesive domains on the parasite surface [11][12][13].
–
As apicomplexan parasites often occupy potentially harmful pro-oxidative ecological niches, such as macrophages or vertebrate erythrocytes, genetic events including horizontal gene transfer might also have shaped the parasites’ peculiar redox metabolism. Peroxiredoxins are central players of the redox metabolism in pro- and eukaryotes. These usually highly concentrated enzymes detoxify hydroperoxides but can also exert functions as temporal redox sensors and chaperones [14][15][16][17][18]. The genome of P. falciparum encodes five different peroxiredoxins [19], some of which have been shown to play a role in parasite development in vivo [20][21]. Furthermore, the parasite imports a highly abundant host peroxiredoxin for hydroperoxide removal [22]. We recently characterized the so-called P. falciparum antioxidant protein (PfAOP) as a model enzyme for the Prx5 subfamily of peroxiredoxins in vitro [23]. PfAOP is rather unusual because of its ability to reduce hydrophobic hydroperoxides with glutathione and glutaredoxin as electron donors [23][24][25][26]. So far, this enzyme has not been characterized in vivo. Here we show that PfAOP is dually localized to the parasite plastid and cytosol. The plastid localization is not the result of a gene acquisition by secondary endosymbiosis but presumably originates from a gene fusion event after a horizontal prokaryote-to-eukaryote gene transfer in a marine apicomplexan ancestor. These findings provide novel insights into the evolution of apicomplexan parasites and peroxiredoxins and might initiate the discovery of overseen horizontal gene transfer events.
RESULTS
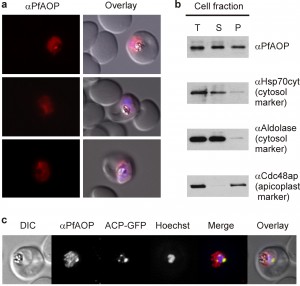
PfAOP is dual localized to the apicoplast and cytosol
PfAOP was previously predicted to localize to the apicoplast [25][26], and an episomally encoded fusion construct between GFP and the N-terminus of PfAOP without its Prx5 domain was indeed detected in this plastid organelle [27]. When we analysed the subcellular localization of endogenous PfAOP by immunofluorescence microscopy, we observed punctate as well as network structures that are characteristic for the apicoplast [28]. Much to our surprise, we furthermore detected a rather strong fluorescence in the parasite cytosol (Fig. 1A). Our purified peptide antibodies used for the localization experiments were specific for PfAOP (Fig. S1), and immunofluorescence microscopy with preimmune serum gave no signal under the same experimental conditions. We therefore hypothesised that PfAOP is dual localized. To further address this possibility, we generated P. falciparum cell extracts by saponin lysis and performed a subcellular fractionation. Subsequent western blot analyses revealed a dual localization of PfAOP in the cytosolic fraction as well as the organellar, apicoplast-containing fraction (Fig. 1B). Cytosolic Hsp70 and aldolase as well as Cdc48 in the apicoplast served as established marker proteins [29]. The apparent size of PfAOP in both subcellular fractions was approximately 22 kDa and therefore slightly smaller than recombinant His-tagged PfAOP∆59 which runs at approximately 25 kDa [23] (Fig. 1B and Fig. S1). Unprocessed full length PfAOP with a calculated molecular mass of 28.1 kDa was not observed (Fig. S1). In addition, the punctate structures in Fig. 1A were confirmed to represent the apicoplast, as revealed by co-localization experiments with transgenic parasites that expressed a GFP-tagged apicoplast marker protein (Fig. 1C). In summary, PfAOP is not exclusively an apicoplast protein but is actually dual localized to the apicoplast and the parasite cytosol.
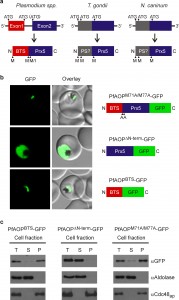
A modular gene architecture reflects the dual localization
The gene architecture of PfAOP is conserved among Plasmodium species: Exon1 encodes the bipartite topogenic signal (BTS) consisting of an ER-type signal followed by a plant-like transit peptide required for targeting to the apicoplast [28]. Please note that the BTS is usually processed upon protein import [28] in accordance with the observed size of mature PfAOP in Fig. 1B and Fig. S1. Exon2 encodes the Prx5 domain (Fig. 2A and Fig. S2). The BTS is necessary and sufficient to target PfAOP to the apicoplast, as revealed by fluorescence microscopy and subcellular fractionation assays with transgenic parasites expressing different GFP-tagged PfAOP constructs. Removal of the N-terminus in PfAOP∆N-term-GFP abrogated the apicoplast targeting, whereas constructs with mutated methionine residues at the start of the Prx5 domain (PfAOPM71A/M77A-GFP) or without the C-terminal Prx5 domain (PfAOPBTS-GFP) were found exclusively in the apicoplast (Fig. 2B,C and Fig. S3). To analyse the evolutionary conservation of the modular architecture of PfAOP and its Plasmodium homologues, we searched the genomes of other apicomplexan parasites for similar genes. We were able to identify Prx5 homologues in Toxoplasma gondii and Neospora caninum but not in Cryptosporidium, Eimeria, Babesia and Theileria. The Prx5 homologues from Toxoplasma and Neospora are encoded by a single exon (Fig. 2A) and have significantly altered N-termini (Fig. S4).
Furthermore, in contrast to their BTS-containing Plasmodium homologues, the Toxoplasma and Neospora proteins are predicted to localize to the mitochondrial matrix or the endoplasmic reticulum (using a variety of prediction programmes as listed in the methods section). In summary, Prx5 homologues are found in several apicomplexan parasites but the modular gene architecture of PfAOP with its separate BTS-encoding exon1 appears to be specific for Plasmodium species.
–
Prokaryotic ancestry and gene fusion of PfAOP
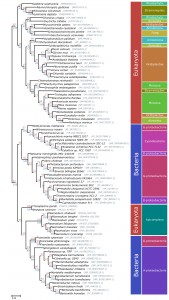
The apicoplast of apicomplexan parasites is a chloroplast-like organelle that was most likely acquired by secondary endosymbiosis after engulfment of a single-celled red alga by an auxotrophic protist [30][31]. As a result, numerous genes of apicomplexan parasites, such as ApiAP2 transcription factors, the histone modifier Ashr3 and fatty acid synthases, probably have an algal origin [4][7][12][13]. Since PfAOP carries a BTS, we wanted to analyse its phylogenetic origin and potential acquisition by secondary endosymbiosis. Although PfAOP and plant Prx5 isoforms share several properties on the protein level [23][24][32], PfAOP and its apicomplexan homologues did not cluster together with algal or plant homologues in phylogenetic sequence analyses (Fig. 3). In contrast, highest similarities were found between apicomplexan and bacterial Prx5 homologues. Even though the bootstrap values in the resulting phylogenetic tree were low, none of the bootstrapped trees recovered the apicomplexan clade as a monophyletic branch with any algae or alveolate species. We further discarded such hypotheses by testing 96 alternative tree topologies with different constrained positions for the monophyletic apicomplexan clade. The results were consistent regardless of the alignment trimming strategy or phylogenetic inference methodology used (i.e. maximum likelihood or Bayesian inference).
Altogether, even though the phylogenetic signal was not sufficiently strong to elucidate a clear ancestor for the PfAOP gene, our tests revealed a phylogenetic incongruence that excludes a horizontal gene transfer from an algal nucleus and that is in agreement with a direct prokaryotic ancestry of PfAOP and its apicomplexan homologues. Hence, the apicoplast localization of PfAOP is not a consequence of secondary endosymbiosis, but is presumably derived from the secondary addition of exon1.
–
Please note that we were unable to identify similar bacterial-like Prx5 homologues in dinoflagellates or other alveolates by BLAST searches. This excludes a mitochondrial ancestry of PfAOP and suggests a horizontal prokaryote-to-eukaryote gene transfer after the divergence of the apicomplexan and dinoflagellate lineages. (The few Prx5 homologues that were identified in other alveolates, including two Perkinsus species and the ciliates Oxytricha trifallax and Ichthyophthirius multifiliis, are in the exclusively eukaryotic branch of Fig. 3, which might point to a horizontal gene transfer resulting from secondary endosymbiosis for these selected genes). In summary, our data support a prokaryotic ancestry and contradict a plastid or mitochondrial origin of apicomplexan Prx5 homologues.
DISCUSSION
When exactly might the prokaryote-to-eukaryote gene transfer of a Prx5 homologue have occurred? Among apicomplexan parasites, the blood parasites Babesia and Theileria are closely related to Plasmodium and are altogether classified as aconoidasida, whereas Cryptosporidium and the coccidia Toxoplasma, Neospora and Eimeria are conoidasida [6]. The presence of similar Prx5 homologues in aconoidasida and conoidasida (Fig. 2A and Fig. 3) suggests that the gene was acquired before both parasite lines have diverged – which presumably occurred before the Cambrian radiation [33] – because a single gene acquisition followed by the loss of the bacterial-like Prx5 homologue in some apicomplexans is much more likely than independent gene acquisitions in Plasmodium, Toxoplasma and Neospora. A plausible evolutionary scenario is therefore a horizontal gene transfer between a marine bacterium and an ancestor of apicomplexans. Such an ancestor might have shared significant similarities with the closely related photosynthetic coral symbionts Chromera velia or Vitrella brassicaformis [30][34], and it will be interesting to analyse whether these organisms possess Prx5 homologues of PfAOP. Corals would actually provide an excellent environment for a prokaryote-to-eukaryote gene transfer because they act as common hosts for bacterial and eukaryotic symbionts. Regarding the bacterial source of the apicomplexan Prx5 homologues, Fig. 3 may suggest an alphaproteobacterium, however, based on the bootstrap values, we cannot exclude another bacterial origin. After horizontal gene transfer, the BTS of the Plasmodium homologues was presumably acquired by the secondary addition of exon1. Such an evolutionary scenario is in agreement with a previous report on exon shuffling as a likely source for apicoplast targeting in Plasmodium [35].
–
As far as the redox metabolism of P. falciparum is concerned, an important implication of the dual localization of PfAOP is that the reducing agents glutathione and cytosolic glutaredoxin are able to interact with PfAOP under physiological conditions in vivo in accordance with our previous mechanistic studies in vitro [23]. Furthermore, because of its dual localization, PfAOP could be a candidate not only for temporal redox sensing, as described previously for other peroxiredoxins [18], but for spatiotemporal redox sensing in different subcellular compartments during the parasite’s sophisticated life cycle.
–
In conclusion, our studies reveal a dual localization of the Prx5 homologue PfAOP in the parasite apicoplast and cytosol. The dual localization is reflected by a modular gene architecture, which appears to be conserved in Plasmodium but not in other apicomplexan parasites. PfAOP was not acquired by secondary endosymbiosis but presumably results from a gene fusion event after a horizontal prokaryote-to-eukaryote gene transfer in a marine apicomplexan ancestor. Hence, PfAOP and related proteins from Toxoplasma and Neospora form a novel subclass of Prx5 homologous with prokaryotic ancestry. In addition, PfAOP reflects an intriguing example for the addition of a gene by horizontal prokaryote-to-eukaryote gene transfer in an otherwise highly economised parasite genome. The physiological relevance and the advantage of targeting PfAOP to the apicoplast remain to be studied. Another aspect noteworthy is that some intracellular alphaproteobacterial or gammaproteobacterial pathogens, such as Bartonella or Francisella species, do not only possess Prx5 homologues that are extremely similar to the apicomplexan homologues, but also share vertebrate host cells and arthropod vectors with apicomplexan parasites. It might therefore be promising to analyse potentially analogous Prx5 functions in these pathogens as well as putative pathogen-to-pathogen gene transfers upon co-infection.
MATERIALS AND METHODS
Cloning of PfAOP-GFP constructs
The full-length pfaop gene was PCR amplified from a cDNA library of P. falciparum strain 3D7 and cloned into the vector pDrive (Qiagen) using the restriction site-containing primers fl_PfAOP/pARL/s (5′-GCGCCCATGGTATAAATGAGAATGAGAAGAACAATAC-3′) and PfAOP/pARL/as (5′-GCGCCCTAGGACCATTACCTAACTGATTATTTTTTAAAAACTCTTTTAC-3′). The truncation constructs pfaop∆Nterm and pfaopBTS were amplified from full-length pfaop using the alternative primers sh_PfAOP/pARL/s (5′-GCGCCCATGGCAAAAGAAAATGATCTTATTCCTAACG-3′) and BTS_PfAOP/pARL/as (5′-GCGCCCTAGGACCATTAGGAATAAGATCATTTTCTTTTATATC-3′), respectively. Potential alternative start codons in full-length pfaop were replaced by site-directed mutagenesis using the primers PfAOP-M71A/M77A/s (5′-CGTGAAAGTTGCAATTGACGTAAGAAATGCAAACAACAT ATC-3′) and PfAOP-M71A/M77A/as (5′-GATATGTTGTTTGCATTTCTTACGTCAATTGCAACTTTCACG-3′). Next, an NcoI restriction site in the GFP-coding sequence of pfgloI-gfp/pARL [36] was removed by site-directed mutagenesis using the primers GFP∆NcoI/s (5′-GAAAACTACCTGTTCCTTGGCCAACACTTGTCAC-3′) and GFP∆NcoI/as (5′-GTGACAAGTGTTGGCCAAGGAACAGGTAGTTTTCC-3′). The gene pfgloI was subsequently excised using AvrII and NcoI, and full length and truncated pfaop was cloned into the vector pARL after digestion with the same restriction enzymes. Correct sequences of all pfaop-gfp/pARL constructs were confirmed by commercial sequencing (MWG Eurofins).
–
P. falciparum culture and transfection
P. falciparum strain 3D7 was cultured at 37°C according to standard protocols [37] using fresh A+ human red blood cells at 3% hematocrit in RPMI 1640 medium that was supplemented with 0.45% (w/v) Albumax II, 200 µM hypoxanthine and 8.9 µg/µl gentamicin. Parasite cultures were maintained at 80% humidity, 5% CO2, 5% O2 and 90% N2. When necessary, parasites were synchronized using the sorbitol method [38]. Transfections were performed as previously described [39] using 100 µg of plasmid DNA per construct. Parasites were subsequently selected with 5 nM WR99210. Transfectants were detected in blood smears between two and four weeks post transfection.
–
Generation and purification of antibodies
Two rabbit peptide antibodies against residues 79-92 and 208-226 of PfAOP were generated (Pineda Antibody Service) and subsequently purified from sera by affinity chromatography using the peptides NH2-CNISDTDGSPNDFTS-CONH2 (αPfAOP-2) and NH2-CMFQEKDKQHNIQTDPYDIS-CONH2 (αPfAOP-5) according to an established protocol [40]. The purity of the eluate fractions was verified by SDS-PAGE and the specificity of the peptide antibodies was assessed by immunoblotting against lysates of both uninfected and infected red blood cells. Preimmune sera served as controls (Fig. S1).
–
Live cell imaging
Live parasites were washed twice with Ringer’s solution (122.5 mm NaCl, 5.4 mm KCl, 1.2 mm CaCl2, 0.8 mm MgCl2, 11 mm d-glucose, 10 mm HEPES, 1 mm NaH2PO4, pH 7.4) and visualized using a LSM510 confocal laser scanning microscope (Zeiss). GFP fluorescence was detected using an argon laser with 3% laser transmission, 40% output, 505-530 nm band pass filter and excitation at 488 nm. All images were processed with the software ImageJ [41] and are representative of at least 20 individual observations for each transgenic parasite line. A Gaussian filter of radius 1.0 was applied on all images. No gamma correction was performed.
–
Immunofluorescence analyses
Indirect immunofluorescence microscopy was carried out as previously described [29] using αPfAOP-2 (1:50) and anti-rabbit Cy3 (1:2000) on either wild-type 3D7 or transgenic ACP-GFP [42] parasites. Preimmune serum at the same dilution gave no signal. Cells were counterstained with Hoechst 33258 (50 ng/ml) to visualise the nucleus. Fixed parasites were imaged on an epifluorescence Zeiss Cell Observer system, z-stack images were subjected to deconvolution (Zeiss Axiovision), maximally projected, converted to 8-bit TIFF format, pseudocoloured and overlayed (ImageJ, version 1.48). All images presented are representative of at least 20 individual observations.
–
Parasite purification and subcellular fractionation
The subcellular fractionation of P. falciparum infected red blood cells was performed as previously described with slight modifications [43]. Briefly, parasites at the trophozoite stage were purified by magnetic cell separation [44] on a VarioMACS column (Miltenyi Biotec) and released from red blood cells using 0.1% saponin in PBS (1.84 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4). Freed parasites were re-suspended in PBS that was supplemented with protease inhibitor cocktail (Roche). Parasites were then lysed by five freeze-thaw cycles. The obtained homogenate, herein referred to as the total parasite lysate, was centrifuged (16,000 g, 60 min, 4°C) to separate the soluble supernatant fraction (S), which represents the cytosolic fraction, from the organelle-enriched pellet fraction (P). After removal of the supernatant, the pellet was washed twice with PBS. The three fractions were then mixed with Laemmli buffer supplemented with 30% 2-mercaptoethanol, boiled for 5 min at 95°C and analysed by SDS-PAGE and western blotting. The following primary antibodies were used at the indicated concentrations: αPfHsp70 (1:1000), αPfCdc48 (1:1000), αPfAldolase (1:5000), αGFP (1:2000), αPfAOP-2 (1:100) and αPfAOP-5 (1:200). Proteins were detected by chemiluminescence using goat anti-mouse HRP-conjugated secondary antibody (1:6000, Bio-Rad) for PfHsp70 and goat anti-rabbit HRP-conjugated secondary antibody (1:10000, Bio-Rad) for all other antibodies.
–
Phylogenetic analyses and bioinformatics
All phylogenetic analyses were carried out using 84 homologous manually selected Prx5 amino acid sequences, covering 44 eukaryotes and 40 bacterial species. The first exon of the Plasmodium sequences was removed prior to all analyses. Multiple sequence alignment was produced using MAFFT v7 [45] with the LINSi algorithm and a gap-opening penalty of 0.1. Evolutionary model selection was performed prior to phylogenetic inference using ProtTest v3 [46] with default parameters, obtaining WAG as the best fitting model. Tree inference was carried out using both maximum likelihood (RAxML v8 [47], rapid hill climbing algorithm, gamma distribution, WAG model, 100 runs and 100 bootstrap replicates) and Bayesian approaches (MrBayes [48], 4 chains, fixed WAG model, gamma distribution, 4 rates categories, 5,000,000 generations). A trimmed version of the alignment, generated with TrimAl v1.3 [49] and gap threshold of 40%, was also evaluated, obtaining no significant differences in the topology and bootstrap supports. The relative position of the apicomplexan proteins was manually inspected for each of the 100 bootstrapped trees, obtaining 96 topologies in which the apicomplexan proteins were still grouped within the bacteria clade, and 4 topologies in which Plasmodium and coccidian species were split, the first being grouped as part of the eukaryotic clade and the former within bacteria. Similarly, all the sub-optimal topologies obtained from 100 RAxML runs were evaluated separately, obtaining no trees in which the apicomplexan proteins were recovered within the eukaryotic clade. Tree hypothesis testing was performed using CONSEL [50] on 96 alternative topologies covering all possible phylogenetic positions of the apicomplexan clade as a monophyletic group with any of the alveolate or algae species considered in the tree (http://github.com/jhcepas/prx5_supplementary_data). For this, 96 constrained topologies were manually generated and subsequently optimized using RAxML with the same parameters described above. All alternative phylogenies were evaluated with CONSEL. The 96 evaluated alternative topologies were rejected with P-values < 0.01 (73), < 0.05 (12) and < 0.1 (11) using the Approximately Unbiased (AU) test [51]. Tree visualization, the generation of constrained tree topologies and the inspection of bootstrapped and sub-optimal tree topologies were performed using ad-hoc scripts based on the ETE toolkit [52]. The complete list of topologies evaluated, source data, software and scripts are available as supplementary data at (http://github.com/jhcepas/prx5_supplementary_data). The subcellular localization of PfAOP and homologues from other alveolates were predicted using a variety of available bioinformatics tools including PATS [53], Predotar [54], PlasMit [55] and Wolf and iPSORT [56].
References
- P.J. Keeling, and J.D. Palmer, "Horizontal gene transfer in eukaryotic evolution", Nature Reviews Genetics, vol. 9, pp. 605-618, 2008. http://dx.doi.org/10.1038/nrg2386
- J.C. Dunning Hotopp, "Horizontal gene transfer between bacteria and animals", Trends in Genetics, vol. 27, pp. 157-163, 2011. http://dx.doi.org/10.1016/j.tig.2011.01.005
- O. Zhaxybayeva, and W.F. Doolittle, "Lateral gene transfer", Current Biology, vol. 21, pp. R242-R246, 2011. http://dx.doi.org/10.1016/j.cub.2011.01.045
- M.J. Gardner, N. Hall, E. Fung, O. White, M. Berriman, R.W. Hyman, J.M. Carlton, A. Pain, K.E. Nelson, S. Bowman, I.T. Paulsen, K. James, J.A. Eisen, K. Rutherford, S.L. Salzberg, A. Craig, S. Kyes, M. Chan, V. Nene, S.J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M.W. Mather, A.B. Vaidya, D.M.A. Martin, A.H. Fairlamb, M.J. Fraunholz, D.S. Roos, S.A. Ralph, G.I. McFadden, L.M. Cummings, G.M. Subramanian, C. Mungall, J.C. Venter, D.J. Carucci, S.L. Hoffman, C. Newbold, R.W. Davis, C.M. Fraser, and B. Barrell, "Genome sequence of the human malaria parasite Plasmodium falciparum", Nature, vol. 419, pp. 498-511, 2002. http://dx.doi.org/10.1038/nature01097
- C. Martens, K. Vandepoele, and Y. Van de Peer, "Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species", Proceedings of the National Academy of Sciences, vol. 105, pp. 3427-3432, 2008. http://dx.doi.org/10.1073/pnas.0712248105
- S.M. Adl, A.G.B. Simpson, C.E. Lane, J. Lukeš, D. Bass, S.S. Bowser, M.W. Brown, F. Burki, M. Dunthorn, V. Hampl, A. Heiss, M. Hoppenrath, E. Lara, L. le Gall, D.H. Lynn, H. McManus, E.A.D. Mitchell, S.E. Mozley‐Stanridge, L.W. Parfrey, J. Pawlowski, S. Rueckert, L. Shadwick, C.L. Schoch, A. Smirnov, and F.W. Spiegel, "The Revised Classification of Eukaryotes", Journal of Eukaryotic Microbiology, vol. 59, pp. 429-514, 2012. http://dx.doi.org/10.1111/j.1550-7408.2012.00644.x
- J. Huang, N. Mullapudi, T. Sicheritz-Ponten, and J.C. Kissinger, "A first glimpse into the pattern and scale of gene transfer in the Apicomplexa", International Journal for Parasitology, vol. 34, pp. 265-274, 2004. http://dx.doi.org/10.1016/j.ijpara.2003.11.025
- B. Striepen, M.W. White, C. Li, M.N. Guerini, S. Malik, J.M. Logsdon, C. Liu, and M.S. Abrahamsen, "Genetic complementation in apicomplexan parasites", Proceedings of the National Academy of Sciences, vol. 99, pp. 6304-6309, 2002. http://dx.doi.org/10.1073/pnas.092525699
- B. Striepen, A.J.P. Pruijssers, J. Huang, C. Li, M. Gubbels, N.N. Umejiego, L. Hedstrom, and J.C. Kissinger, "Gene transfer in the evolution of parasite nucleotide biosynthesis", Proceedings of the National Academy of Sciences, vol. 101, pp. 3154-3159, 2004. http://dx.doi.org/10.1073/pnas.0304686101
- J. Huang, N. Mullapudi, C.A. Lancto, M. Scott, M.S. Abrahamsen, and J.C. Kissinger, "Array", Genome Biology, vol. 5, pp. R88, 2004. http://dx.doi.org/10.1186/gb-2004-5-11-r88
- T.J. Templeton, "Whole-genome natural histories of apicomplexan surface proteins", Trends in Parasitology, vol. 23, pp. 205-212, 2007. http://dx.doi.org/10.1016/j.pt.2007.03.001
- S.P. Kishore, J.W. Stiller, and K.W. Deitsch, "Horizontal gene transfer of epigenetic machinery and evolution of parasitism in the malaria parasite Plasmodium falciparum and other apicomplexans", BMC Evolutionary Biology, vol. 13, 2013. http://dx.doi.org/10.1186/1471-2148-13-37
- S. Balaji, "Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains", Nucleic Acids Research, vol. 33, pp. 3994-4006, 2005. http://dx.doi.org/10.1093/nar/gki709
- H.H. Jang, K.O. Lee, Y.H. Chi, B.G. Jung, S.K. Park, J.H. Park, J.R. Lee, S.S. Lee, J.C. Moon, J.W. Yun, Y.O. Choi, W.Y. Kim, J.S. Kang, G. Cheong, D. Yun, S.G. Rhee, M.J. Cho, and S.Y. Lee, "Two Enzymes in One", Cell, vol. 117, pp. 625-635, 2004. http://dx.doi.org/10.1016/j.cell.2004.05.002
- R. Brigelius-Flohé, and L. Flohé, "Basic Principles and Emerging Concepts in the Redox Control of Transcription Factors", Antioxidants & Redox Signaling, vol. 15, pp. 2335-2381, 2011. http://dx.doi.org/10.1089/ars.2010.3534
- A. Hall, K. Nelson, L.B. Poole, and P.A. Karplus, "Structure-based Insights into the Catalytic Power and Conformational Dexterity of Peroxiredoxins", Antioxidants & Redox Signaling, vol. 15, pp. 795-815, 2011. http://dx.doi.org/10.1089/ars.2010.3624
- S.G. Rhee, and H.A. Woo, "Multiple Functions of Peroxiredoxins: Peroxidases, Sensors and Regulators of the Intracellular Messenger H2O2, and Protein Chaperones", Antioxidants & Redox Signaling, vol. 15, pp. 781-794, 2011. http://dx.doi.org/10.1089/ars.2010.3393
- R.S. Edgar, E.W. Green, Y. Zhao, G. van Ooijen, M. Olmedo, X. Qin, Y. Xu, M. Pan, U.K. Valekunja, K.A. Feeney, E.S. Maywood, M.H. Hastings, N.S. Baliga, M. Merrow, A.J. Millar, C.H. Johnson, C.P. Kyriacou, J.S. O’Neill, and A.B. Reddy, "Peroxiredoxins are conserved markers of circadian rhythms", Nature, vol. 485, pp. 459-464, 2012. http://dx.doi.org/10.1038/nature11088
- M.C. Gretes, L.B. Poole, and P.A. Karplus, "Peroxiredoxins in Parasites", Antioxidants & Redox Signaling, vol. 17, pp. 608-633, 2012. http://dx.doi.org/10.1089/ars.2011.4404
- D. Richard, R. Bartfai, J. Volz, S.A. Ralph, S. Muller, H.G. Stunnenberg, and A.F. Cowman, "A Genome-wide Chromatin-associated Nuclear Peroxiredoxin from the Malaria Parasite Plasmodium falciparum", Journal of Biological Chemistry, vol. 286, pp. 11746-11755, 2011. http://dx.doi.org/10.1074/jbc.M110.198499
- B.A. Turturice, M.A. Lamm, J.J. Tasch, A. Zalewski, R. Kooistra, E.H. Schroeter, S. Sharma, S. Kawazu, and S.M. Kanzok, "Expression of Cytosolic Peroxiredoxins in Plasmodium berghei Ookinetes Is Regulated by Environmental Factors in the Mosquito Bloodmeal", PLoS Pathogens, vol. 9, pp. e1003136, 2013. http://dx.doi.org/10.1371/journal.ppat.1003136
- S. Koncarevic, P. Rohrbach, M. Deponte, G. Krohne, J.H. Prieto, J. Yates, S. Rahlfs, and K. Becker, "The malarial parasite Plasmodium falciparum imports the human protein peroxiredoxin 2 for peroxide detoxification", Proceedings of the National Academy of Sciences, vol. 106, pp. 13323-13328, 2009. http://dx.doi.org/10.1073/pnas.0905387106
- C.F. Djuika, S. Fiedler, M. Schnölzer, C. Sanchez, M. Lanzer, and M. Deponte, "Plasmodium falciparum antioxidant protein as a model enzyme for a special class of glutaredoxin/glutathione-dependent peroxiredoxins", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1830, pp. 4073-4090, 2013. http://dx.doi.org/10.1016/j.bbagen.2013.04.020
- M. Deponte, "Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes", Biochimica et Biophysica Acta (BBA) - General Subjects, vol. 1830, pp. 3217-3266, 2013. http://dx.doi.org/10.1016/j.bbagen.2012.09.018
- C. Nickel, S. Rahlfs, M. Deponte, S. Koncarevic, and K. Becker, "Thioredoxin Networks in the Malarial ParasitePlasmodium falciparum", Antioxidants & Redox Signaling, vol. 8, pp. 1227-1239, 2006. http://dx.doi.org/10.1089/ars.2006.8.1227
- M. Deponte, S. Rahlfs, and K. Becker, "Peroxiredoxin systems of protozoal parasites.", Sub-cellular biochemistry, 2007. http://www.ncbi.nlm.nih.gov/pubmed/18084896
- S. Kehr, N. Sturm, S. Rahlfs, J.M. Przyborski, and K. Becker, "Compartmentation of Redox Metabolism in Malaria Parasites", PLoS Pathogens, vol. 6, pp. e1001242, 2010. http://dx.doi.org/10.1371/journal.ppat.1001242
- R.F. Waller, "Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway", The EMBO Journal, vol. 19, pp. 1794-1802, 2000. http://dx.doi.org/10.1093/emboj/19.8.1794
- S. Spork, J.A. Hiss, K. Mandel, M. Sommer, T.W.A. Kooij, T. Chu, G. Schneider, U.G. Maier, and J.M. Przyborski, "An Unusual ERAD-Like Complex Is Targeted to the Apicoplast ofPlasmodium falciparum", Eukaryotic Cell, vol. 8, pp. 1134-1145, 2009. http://dx.doi.org/10.1128/EC.00083-09
- J. Janouškovec, A. Horák, M. Oborník, J. Lukeš, and P.J. Keeling, "A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids", Proceedings of the National Academy of Sciences, vol. 107, pp. 10949-10954, 2010. http://dx.doi.org/10.1073/pnas.1003335107
- G.G. van Dooren, and B. Striepen, "The Algal Past and Parasite Present of the Apicoplast", Annual Review of Microbiology, vol. 67, pp. 271-289, 2013. http://dx.doi.org/10.1146/annurev-micro-092412-155741
- K. Dietz, "Peroxiredoxins in Plants and Cyanobacteria", Antioxidants & Redox Signaling, vol. 15, pp. 1129-1159, 2011. http://dx.doi.org/10.1089/ars.2010.3657
- A.A. Escalante, and F.J. Ayala, "Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes.", Proceedings of the National Academy of Sciences of the United States of America, 1995. http://www.ncbi.nlm.nih.gov/pubmed/7597031
- M. Oborník, D. Modrý, M. Lukeš, E. Černotíková-Stříbrná, J. Cihlář, M. Tesařová, E. Kotabová, M. Vancová, O. Prášil, and J. Lukeš, "Morphology, Ultrastructure and Life Cycle of Vitrella brassicaformis n. sp., n. gen., a Novel Chromerid from the Great Barrier Reef", Protist, vol. 163, pp. 306-323, 2012. http://dx.doi.org/10.1016/j.protis.2011.09.001
- C.J. Tonkin, B.J. Foth, S.A. Ralph, N. Struck, A.F. Cowman, and G.I. McFadden, "Evolution of malaria parasite plastid targeting sequences", Proceedings of the National Academy of Sciences, vol. 105, pp. 4781-4785, 2008. http://dx.doi.org/10.1073/pnas.0707827105
- M. Urscher, J.M. Przyborski, M. Imoto, and M. Deponte, "Distinct subcellular localization in the cytosol and apicoplast, unexpected dimerization and inhibition of Plasmodium falciparum glyoxalases", Molecular Microbiology, vol. 76, pp. 92-103, 2010. http://dx.doi.org/10.1111/j.1365-2958.2010.07082.x
- W. Trager, and J.B. Jensen, "Human malaria parasites in continuous culture.", Science (New York, N.Y.), 1976. http://www.ncbi.nlm.nih.gov/pubmed/781840
- C. Lambros, and J.P. Vanderberg, "Synchronization of Plasmodium falciparum erythrocytic stages in culture.", The Journal of parasitology, 1979. http://www.ncbi.nlm.nih.gov/pubmed/383936
- K. Deitsch, C. Driskill, and T. Wellems, "Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes.", Nucleic acids research, 2001. http://www.ncbi.nlm.nih.gov/pubmed/11160909
- E. Eckers, M. Cyrklaff, L. Simpson, and M. Deponte, "Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries", bchm, vol. 393, pp. 513-524, 2012. http://dx.doi.org/10.1515/hsz-2011-0255
- C.A. Schneider, W.S. Rasband, and K.W. Eliceiri, "NIH Image to ImageJ: 25 years of image analysis.", Nature methods, 2012. http://www.ncbi.nlm.nih.gov/pubmed/22930834
- R.F. Waller, P.J. Keeling, R.G. Donald, B. Striepen, E. Handman, N. Lang-Unnasch, A.F. Cowman, G.S. Besra, D.S. Roos, and G.I. McFadden, "Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum.", Proceedings of the National Academy of Sciences of the United States of America, 1998. http://www.ncbi.nlm.nih.gov/pubmed/9770490
- P.M. Southworth, J.E. Hyde, and P.F. Sims, "A mass spectrometric strategy for absolute quantification of Plasmodium falciparum proteins of low abundance", Malaria Journal, vol. 10, 2011. http://dx.doi.org/10.1186/1475-2875-10-315
- T. Staalsoe, H.A. Giha, D. Dodoo, T.G. Theander, and L. Hviid, "Detection of antibodies to variant antigens on Plasmodium falciparum-infected erythrocytes by flow cytometry.", Cytometry, 1999. http://www.ncbi.nlm.nih.gov/pubmed/10213198
- K. Katoh, and H. Toh, "Recent developments in the MAFFT multiple sequence alignment program", Briefings in Bioinformatics, vol. 9, pp. 286-298, 2008. http://dx.doi.org/10.1093/bib/bbn013
- D. Darriba, G.L. Taboada, R. Doallo, and D. Posada, "ProtTest 3: fast selection of best-fit models of protein evolution", Bioinformatics, vol. 27, pp. 1164-1165, 2011. http://dx.doi.org/10.1093/bioinformatics/btr088
- A. Stamatakis, "RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies", Bioinformatics, vol. 30, pp. 1312-1313, 2014. http://dx.doi.org/10.1093/bioinformatics/btu033
- F. Ronquist, M. Teslenko, P. van der Mark, D.L. Ayres, A. Darling, S. Höhna, B. Larget, L. Liu, M.A. Suchard, and J.P. Huelsenbeck, "MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space", Systematic Biology, vol. 61, pp. 539-542, 2012. http://dx.doi.org/10.1093/sysbio/sys029
- S. Capella-Gutiérrez, J.M. Silla-Martínez, and T. Gabaldón, "trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses", Bioinformatics, vol. 25, pp. 1972-1973, 2009. http://dx.doi.org/10.1093/bioinformatics/btp348
- H. Shimodaira, and M. Hasegawa, "CONSEL: for assessing the confidence of phylogenetic tree selection.", Bioinformatics (Oxford, England), 2001. http://www.ncbi.nlm.nih.gov/pubmed/11751242
- H. Shimodaira, "An Approximately Unbiased Test of Phylogenetic Tree Selection", Systematic Biology, vol. 51, pp. 492-508, 2002. http://dx.doi.org/10.1080/10635150290069913
- J. Huerta-Cepas, J. Dopazo, and T. Gabaldón, "ETE: a python Environment for Tree Exploration", BMC Bioinformatics, vol. 11, 2010. http://dx.doi.org/10.1186/1471-2105-11-24
- J. Zuegge, S. Ralph, M. Schmuker, G.I. McFadden, and G. Schneider, "Deciphering apicoplast targeting signals--feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins.", Gene, 2001. http://www.ncbi.nlm.nih.gov/pubmed/11738814
- I. Small, N. Peeters, F. Legeai, and C. Lurin, "Predotar: A tool for rapidly screening proteomes for N‐terminal targeting sequences", PROTEOMICS, vol. 4, pp. 1581-1590, 2004. http://dx.doi.org/10.1002/pmic.200300776
- A. Bender, G.G. van Dooren, S.A. Ralph, G.I. McFadden, and G. Schneider, "Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum.", Molecular and biochemical parasitology, 2003. http://www.ncbi.nlm.nih.gov/pubmed/14599665
- H. Bannai, Y. Tamada, O. Maruyama, K. Nakai, and S. Miyano, "Extensive feature detection of N-terminal protein sorting signals.", Bioinformatics (Oxford, England), 2002. http://www.ncbi.nlm.nih.gov/pubmed/11847077
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We thank Andreas Reichert for providing the αGFP antibody. This work was supported by the Deutsche Forschungsgemeinschaft Grant DE 1431/8-1 (to M. D.).
COPYRIGHT
© 2015

Prokaryotic ancestry and gene fusion of a dual localized peroxiredoxin in malaria parasites by Carine F. Djuika et al. is licensed under a Creative Commons Attribution 4.0 International License.