
Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.

Luminal acetylation of microtubules is not essential for Plasmodium berghei and Toxoplasma gondii survival
Acetylation of α-tubulin at lysine 40 is not essential for cytoskeletal stability in Plasmodium berghei or Toxoplasma gondii, suggesting redundancy and plasticity in microtubule regulation in these parasites.

The dual-site agonist for human M2 muscarinic receptors Iper-8-naphtalimide induces mitochondrial dysfunction in Saccharomyces cerevisiae
S. cerevisiae is a model to study human GPCRs. N-8-Iper, active against glioblastoma via M2 receptor, causes mitochondrial damage in yeast by binding Ste2, highlighting evolutionary conservation of GPCRs.

Integrative Omics reveals changes in the cellular landscape of peroxisome-deficient pex3 yeast cells
To uncover the consequences of peroxisome deficiency, we compared Saccharomyces cerevisiae wild-type with pex3 cells, which lack peroxisomes, employing quantitative proteomics and transcriptomics technologies.

Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
Rebekkah E. Pope1, Patrick Ballmann2, Lisa Whitworth3 and Rolf A. Prade1,*
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
Evelyn Tevere1,a, María G. Mediavilla1,a, Cecilia B. Di Capua1, Marcelo L. Merli1, Carlos Robello2,3, Luisa Berná2,4 and Julia A. Cricco
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.
Sir2 regulates selective autophagy in stationary-phase yeast cells
Ji-In Ryua, Juhye Junga, and Jeong-Yoon Kim
This study establishes Sir2 as a previously unrecognized regulator of selective autophagy during the stationary phase and highlight how cells dynamically control organelle degradation.
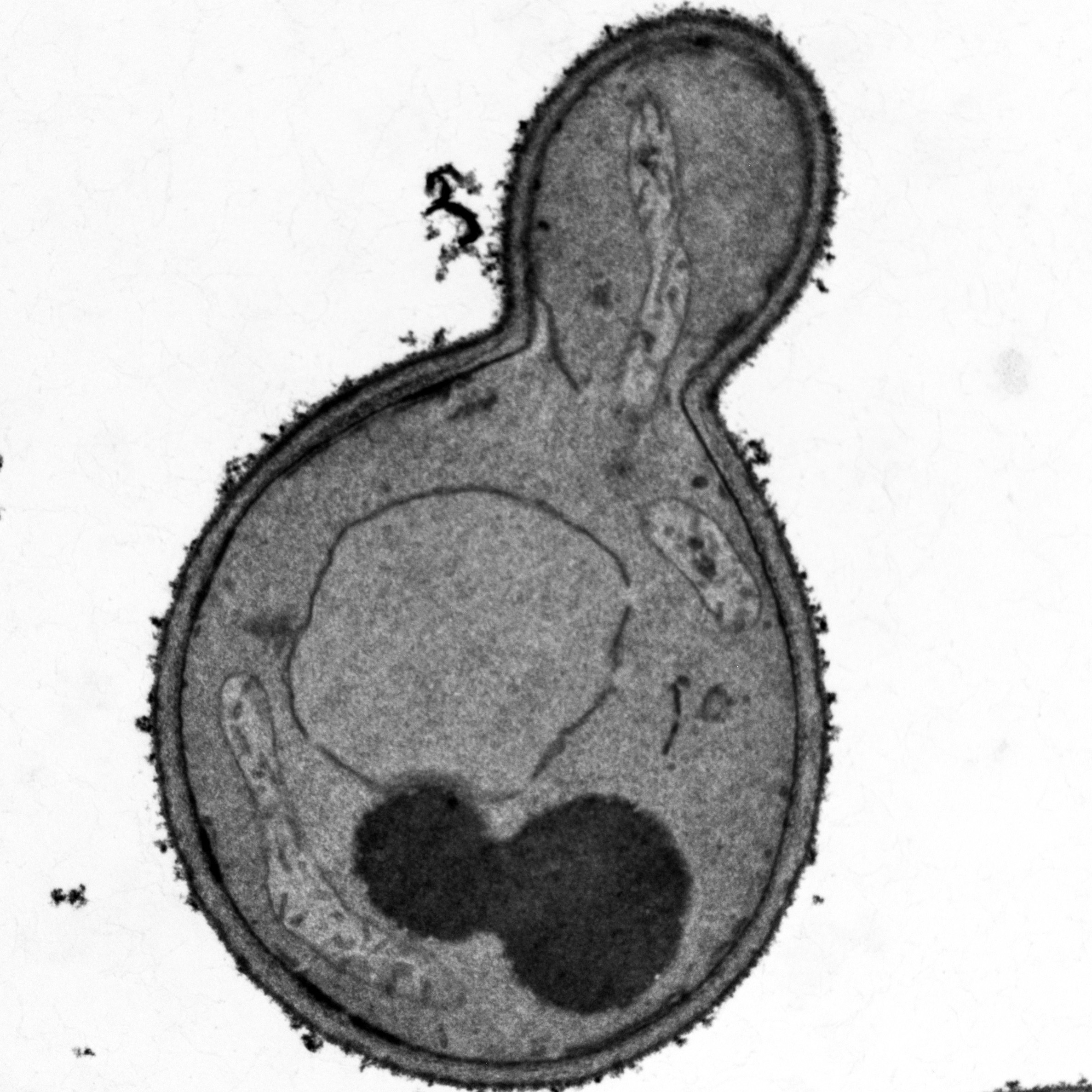
Microwave-assisted preparation of yeast cells for ultrastructural analysis by electron microscopy
Moritz Mayera, Christina Schuga, Stefan Geimer, Till Klecker and Benedikt Westermann
Budding yeast Saccharomyces cerevisiae is widely used as a model organism to study the biogenesis and architecture of organellar membranes, which can be visualized by transmission electron microscopy (TEM).

A complex remodeling of cellular homeostasis distinguishes RSV/SARS-CoV-2 co-infected A549-hACE2 expressing cell lines
Claudia Vanetti1, Irma Saulle1,2, Valentina Artusa1,2, Claudia Moscheni1, Gioia Cappelletti1, Silvia Zecchini1, Sergio Strizzi1, Micaela Garziano1,2, Claudio Fenizia1,2, Antonella Tosoni1, Martina Broggiato1, Pasquale Ogno1, Manuela Nebuloni1, Mario Clerici2,3, Daria Trabattoni1, Fiona Limanaqi1 and Mara Biasin1
Given the common tropism of SARS-CoV-2 and RSV, and the unclear consequences of their mutual influence, we developed an in vitro lung epithelial cell model to study the molecular mechanisms and cellular pathways modulated in viral co-infection.

Fecal gelatinase does not predict mortality in patients with alcohol-associated hepatitis
Yongqiang Yang1,a, Philipp Hartmann2,3,a and Bernd Schnabl1,4
This study aimed to investigate the significance of fecal gelatinase on clinical outcomes in patients with alcohol-associated hepatitis. In conclusion, in our cohort, fecal gelatinase does not predict mortality and does not indicate higher disease severity in patients with alcohol-associated hepatitis.

Direct detection of stringent alarmones (pp)pGpp using malachite green
Muriel Schicketanz1, Magdalena Petrová2, Dominik Rejman2, Margherita Sosio3, Stefano Donadio3 and Yong Everett Zhang1
In this study, we demonstrate the surprising discovery of a commercially available, low-cost malachite green (MG) detection kit, originally designed for orthophosphate (Pi) detection, for detecting (p)ppGpp and its analogues, especially pGpp

Promoter methylation and increased expression of PD-L1 in patients with active tuberculosis
Yen-Han Tseng1,2, Sheng-Wei Pan1,2,3, Jhong-Ru Huang2,4, Chang-Ching Lee1, Jung-Jyh Hung2,5, Po-Kuei Hsu2,5, Nien-Jung Chen6, Wei-Juin Su2,7, Yuh-Min Chen1,2 and Jia-Yih Feng1,2,8
The PD-1/PD-L1 pathway plays a pivotal role in T cell activity and is involved in the pathophysiology of tuberculosis. Here we show that PD-L1 expression is increased in patients with active tuberculosis and is correlated with treatment outcomes.

Quantification methods of Candida albicans are independent irrespective of fungal morphology
Amanda B Soares1, Maria C de Albuquerque1, Leticia M Rosa1, Marlise I Klein 2, Ana C Paravina1, Paula A Barbugli1, Livia N Dovigo3 and Ewerton G de O Mima1
Our study demonstrated that the quantification methods of C. albicans (cells/mL, CFU/mL, and vPCR) did not agree, regardless of the fungal morphology/growth, even though a significant and strong correlation is observed.

Pathogenic Escherichia coli change the adhesion between neutrophils and endotheliocytes in the experimental bacteremia model
Svetlana N Pleskova1,2,*, Nikolay A Bezrukov1, Sergey Z Bobyk1, Ekaterina N Gorshkova1 and Dimitri V Novikov3
In this work, we have demonstrated that in the model of experimental septicemia there is a disruption of adhesion contacts between neutrophils and endothelial cells, manifested by a decrease in adhesion force and work upon exposure to E. coli.

Arsenite treatment induces Hsp90 aggregates distinct from conventional stress granules in fission yeast
Naofumi Tomimotoa, Teruaki Takasakia and Reiko Sugiura
Given the conserved role of Hsp90 as a molecular chaperone protein, our findings presented in this study may suggest a novel type of arsenite-induced biological condensates, wherein Hsp90 plays a key role in maintaining its integrity.
From the Uncharacterized Protein Family 0016 to the GDT1 family: Molecular insights into a newly-characterized family of cation secondary transporters
Louise Thines1, Jiri Stribny1 and Pierre Morsomme1
This review outlines how the formerly uncharacterized UPF0016 family, now known as the Gdt1 family, plays key roles in cation transport – especially Mn²⁺ – across species from bacteria to humans. These proteins are crucial for processes like glycosylation, photosynthesis, and calcium signaling, with functions linked to their localization in membranes such as the Golgi, chloroplast, and plasma membrane and by that highlighting their evolutionary conservation and physiological relevance, offering insights into their shared and distinct features across organisms.
A broad-spectrum antibiotic adjuvant SLAP-S25: one stone many birds
Meirong Song1 and Kui Zhu1
This article refers to the study “A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens” by Song et al. (Nat Microbiol, 2020), which deals with the growing threat of antibiotic resistance, with few new drugs being developed for decades. The study found that the peptide SLAP-S25 enhances the efficacy of several antibiotics against resistant Gram-negative bacteria by disrupting their membranes, thereby increasing drug uptake. This suggests that bacterial membranes are promising targets for new antibiotic adjuvants.
Hiding in plain sight: vesicle-mediated export and transmission of prion-like proteins
Mehdi Kabani1
This article relates to the study “Glucose availability dictates the export of the soluble and prion forms of Sup35p via periplasmic or extracellular vesicles” by Kabani et al. (Mol Microbiol, 2020) that provides compelling evidence that yeast prions, such as Sup35p in its infectious [PSI⁺] state, can be exported via both extracellular vesicles (EVs) and periplasmic vesicles (PVs), with this export being modulated by environmental glucose levels. The discovery that prion particles are released in high amounts through PVs during glucose starvation adds a new dimension to our understanding of prion transmission and opens up fascinating possibilities for exploring vesicle-mediated spread of protein aggregates in neurodegenerative diseases using yeast as a model system.
Regulation of Cdc42 for polarized growth in budding yeast
Kristi E. Miller1,2, Pil Jung Kang1 and Hay-Oak Park1
This review highlights how studies in budding yeast have revealed a biphasic mechanism of Cdc42 activation that governs cell polarity establishment, with implications for understanding similar processes in mammalian cells and the role of Cdc42 in aging.
Yeast-based assays for the functional characterization of cancer-associated variants of human DNA repair genes
Tiziana Cervelli1, Samuele Lodovichi1, Francesca Bellè1 and Alvaro Galli1
This article highlights how the genetic tractability and conserved DNA repair pathways of yeast make it a powerful system for functionally characterizing human cancer-associated variants in DNA repair genes, aiding in risk assessment and therapeutic decision-making.
A novel c-di-GMP signal system regulates biofilm formation in Pseudomonas aeruginosa
Gukui Chen1 and Haihua Liang1
This article relates to the study “The SiaA/B/C/D signaling network regulates biofilm formation in Pseudomonas aeruginosa” by Chen et al. (EMBO J, 2020) that reveals a novel signaling network encoded by the siaABCD operon in Pseudomonas aeruginosa that regulates biofilm and aggregate formation by controlling the diguanylate cyclase activity of SiaD through phosphorylation-dependent interactions with SiaC, highlighting a potential antimicrobial target.
Regulation of anti-microbial autophagy by factors of the complement system
Christophe Viret1, Aurore Rozières1, Rémi Duclaux-Loras1, Gilles Boschetti1, Stéphane Nancey1 and
Mathias Faure1,2
This review explores emerging evidence that components of the complement system, beyond their traditional immune roles, modulate autophagy – particularly xenophagy – thereby influencing cell-autonomous antimicrobial responses during host-pathogen interactions.
More than flipping the lid: Cdc50 contributes to echinocandin resistance by regulating calcium homeostasis in Cryptococcus neoformans
Chengjun Cao1 and Chaoyang Xue1,2
In this article, the authors comment on the study “A mechanosensitive channel governs lipid flippase-mediated echinocandin resistance in Cryptococcus neoformans” by Cao et al. (mBio, 2019), which uncovers a dual role for the lipid flippase subunit Cdc50 in Cryptococcus neoformans, linking lipid translocation and calcium signaling via its interaction with the mechanosensitive channel Crm1, thereby contributing to innate resistance against the antifungal drug caspofungin.
Non-genetic impact factors on chronological lifespan and stress resistance of baker’s yeast
Michael Sauer and Diethard Mattanovich
This article comments on work published by Bisschops et al. (Microbial Cell, 2015), which illustrates how important the choice of the experimental setup is and how culture conditions influcence cellular aging and survival in biotechnological processes.
What’s old is new again: yeast mutant screens in the era of pooled segregant analysis by genome sequencing
Chris Curtin and Toni Cordente
This article comments on work published by Den Abt et al. (Microbial Cell, 2016), which identified genes involved in ethyl acetate formation in a yeast mutant screen based on a new approach combining repeated rounds of chemical mutagenesis and pooled segregant analysis by whole genome sequencing.
The complexities of bacterial-fungal interactions in the mammalian gastrointestinal tract
Eduardo Lopez-Medina1 and Andrew Y. Koh2
This article comments on work published by Lopez-Medina et al. (PLoS Pathog, 2015) and Fan et al. (Nat Med, 2015), which utilize an “artificial” niche, the antibiotic-treated gut with concomitant pathogenic microbe expansion, to gain insight in bacterial-fungal interactions in clinically common scenarios.
Gearing up for survival – HSP-containing granules accumulate in quiescent cells and promote survival
Ruofan Yu and Weiwei Dang
This article comments on work published by Lee et al. (Microbial Cell, 2016), which reports that distinct granules are formed in quiescent and non-quiescent cells, which determines their respective cell fates.
Yeast screening platform identifies FDA-approved drugs that reduce Aβ oligomerization
Triana Amen1,2 and Daniel Kaganovich1
This article comments on work published by Park et al. (Microbial Cell, 2016), which discovered a number of small molecules capable of modulating Aβ aggregation in a yeast model.
Groupthink: chromosomal clustering during transcriptional memory
Kevin A. Morano
In this article, the authors comment on the study “NO1 transcriptional memory leads to DNA zip code-dependent interchromosomal clustering.” by Brickner et al. (Microbial Cell, 2015), discussing the importance and molecular mechanisms of chromosomal clustering during transcriptional memory.
Yeast proteinopathy models: a robust tool for deciphering the basis of neurodegeneration
Amit Shrestha1, 2 and Lynn A. Megeney1, 2, 3
Protein quality control or proteostasis is an essential determinant of basic cell health and aging. Eukaryotic cells have evolved a number of proteostatic mechanisms to ensure that proteins retain functional conformation, or are rapidly degraded when proteins misfold or self-aggregate. This article discusses the use of budding yeast as a robust proxy to study the intersection between proteostasis and neurodegenerative disease.
Microbial Cell
is an open-access, peer-reviewed journal that publishes exceptionally relevant research works that implement the use of unicellular organisms (and multicellular microorganisms) to understand cellular responses to internal and external stimuli and/or human diseases.
you can trust
Can’t find what you’re looking for?
You can browse all our issues and published articles here.
FAQs
Peer-reviewed, open-access research using unicellular organisms (and multicellular microorganisms) to understand cellular responses and human disease.
The journal (founded in 2014) is led by its Editors-in-Chief Frank Madeo, Didac Carmona-Gutierrez, and Guido Kroemer
Microbial Cell has been publishing original scientific literature since 2014, and from the very beginning has been managed by active scientists through an independent Publishing House (Shared science Publishers). The journal was conceived as a platform to acknowledge the importance of unicellular organisms, both as model systems as well as in the biological context of human health and disease.
Ever since, Microbial Cell has very positively developed and strongly grown into a respected journal in the unicellular research community and even beyond. This scientific impact is reflected in the yearly number of citations obtained by articles published in Microbial Cell, as recorded by the Web of Science (Clarivate, formerly Thomson/Reuters):

The scientific impact of Microbial Cell is also mirrored in a series of milestones:
2015: Microbial Cell is included in the Emerging Sources Citation Index (ESCI), a selection of developing journals drafted by Clarivate Analytics based on the candidate’s publishing standards, quality, editorial content, and citation data. Note: As an ESCI-selected journal, Microbial Cell is currently being evaluated in a rigorous and long process to determine an inclusion in the Science Citation Index Expanded (SCIE), which allows the official calculation of Clarivate Analytics’ impact factor.
2016: Microbial Cell is awarded the so-called DOAJ Seal by the selective Directory of Open Access Journals (DOAJ). The DOAJ Seal is an exclusive mark of certification for open access journals granted by DOAJ to journals that adhere to outstanding best practice and achieve an extra high and clear commitment to open access and high publishing standards.
2017: Microbial Cell is included in Pubmed Central (PMC), allowing the archiving of all the journal’s articles in PMC and PubMed.
2019: Microbial Cell is indexed in the prestigious abstract and citation database Scopus after a thorough selection process. This also means that Microbial Cell obtains, for the first time, an official Scopus CiteScore as well as an official journal ranking in the Scimago Journal and Country Ranking.
2022: Microbial Cell’s CiteScore reaches a value of 7.2 for the year 2021, positioning Microbial Cell among the top microbiology journals (previously available CiteScores: 2019: 5.4; 2020: 5.1).
2022: Microbial Cell is indexed in the highly selective Science Citation Index Expanded™, which covers approx. 9,500 of the world’s most impactful journals across 178 scientific disciplines. In their journal selection and curation process, Clarivate´s editors apply 24 ‘quality’ criteria and four ‘impact’ criteria to select the most influential journals in their respective fields. This selection is also a pre-requisite for inclusion in the JCR, which features the impact factor.
2022: Microbial Cell is listed in the Journal Citation Reports™ (JCR), and obtains its first official Journal Impact Factor™ (JIF) for the year 2021: 5.316.Check Article Types and Manuscript Preparation guidelines. Submit online via Scholastica.


Similar environments but diverse fates: Responses of budding yeast to nutrient deprivation.
Saul M. Honigberg
Diploid budding yeast (Saccharomyces cerevisiae) can adopt one of several alternative differentiation fates in response to nutrient limitation, and each of these fates provides distinct biological functions. When different strain backgrounds are taken into account, these various fates occur in response to similar environmental cues, are regulated by the same signal transduction pathways, and share many of the same master regulators. I propose that the relationships between fate choice, environmental cues and signaling pathways are not Boolean, but involve graded levels of signals, pathway activation and master-regulator activity.