Research Articles:
Microbial Cell, Vol. 9, No. 8, pp. 145 - 157; doi: 10.15698/mic2022.08.783
Up-regulation of Osh6 boosts an anti-aging membrane trafficking pathway toward vacuoles
1 Department of Biology, University of Arkansas, Little Rock, AR 72204, USA.
Keywords: Osh6, PI4P, Golgi, vacuole, longevity.
Abbreviations:
ER – endoplasmic reticulum,
LE – late endosome,
PI4P – phosphatidylinositol-4-phosphate,
PM – plasma membrane,
PS – phosphatidylserine,
TGN – trans-Golgi network,
TORC – target of rapamycin complex.
Received originally: 13/08/2021 Received in revised form: 09/06/2022
Accepted: 18/06/2022
Published: 15/07/2022
Correspondence:
Fusheng Tang, Department of Biology, FH 406, University of Arkansas at Little Rock, 2801 S. University Ave., Little Rock, AR 72204-1099; Tel: 501-569-3270; Fax: 501-569-3271; fxtang@ualr.edu
Conflict of interest statement: The authors declare that they have no conflict of interest.
Please cite this article as: Ilham Kadhim, Nazneen Begum, William King, Licheng Xu and Fusheng Tang (2022). Up-regulation of Osh6 boosts an anti-aging membrane trafficking pathway toward vacuoles. Microbial Cell 9(8): 145-157. doi: 10.15698/mic2022.08.783
Abstract
Members of the family of oxysterol-binding proteins mediate non-vesicular lipid transport between membranes and contribute to longevity in different manners. We previously found that a 2-fold up-regulation of Osh6, one of seven yeast oxysterol-binding proteins, remedies vacuolar morphology defects in mid-aged cells, partly down-regulates the target of rapamycin complex 1 (TORC1), and increases the replicative lifespan. At the molecular level, Osh6 transports phosphatidylserine (PS) and phosphatidylinositol-4-phosphate (PI4P) between the endoplasmic reticulum (ER) and the plasma membrane (PM). To decipher how an ER-PM working protein controls vacuolar morphology, we tested genetic interactions between OSH6 and DRS2, whose protein flips PS from the lumen to the cytosolic side of the Golgi, the organelle between ER and vacuoles in many pathways. Up-regulated OSH6 complemented vacuolar morphology of drs2∆ and enriched PI4P on the Golgi, indicating that Osh6 also works on the Golgi. This altered PI4P-enrichment led to a delay in the secretion of the proton ATPase Pma1 to the PM and a rerouting of Pma1 to vacuoles in a manner dependent on the trans-Golgi network (TGN) to late endosome (LE) trafficking pathway. Since the TGN-LE pathway controls endosomal and vacuolar TORC1, it may be the anti-aging pathway boosted by up-regulated Osh6.
INTRODUCTION
The family of oxysterol-binding proteins is conserved in all eukaryotes. They mediate non-vesicular lipid transport between membranes and thus play critical roles in cell growth and development [1]. Knocking out all seven oxysterol-binding proteins encoded by OSH genes in yeast leads to highly fragmented vacuoles and cell death [2]. Late endosomes, the organelle immediately upstream of vacuoles (mammalian lysosomes) in multiple membrane trafficking pathways, are also targets of oxysterol-binding proteins. In Caenorhabditis elegans, knocking down the expression of all four oxysterol-binding proteins leads to enlarged late endosome (LE) [3]. In HeLa cells, knocking down the expression of oxysterol-binding protein ORP1L leads to enlarged LE [3]. Since enlarged LE is usually a sign of senescent cells [4], normal functions of oxysterol-binding proteins are thus critical for longevity.
–
We previously found that a 2-fold up-regulation of Osh6 (PERG6–OSH6) remedies vacuolar morphology defects in mid-aged cells and extends the replicative lifespan. Interestingly, the level of Osh6 protein in wild type cells declines with age. Moreover, up-regulation of Osh6 partly represses the target of rapamycin complex 1 (TORC1) but requires TORC1 for longevity [5]. This two-way relationship with TORC1 is supported by a recent study on the spatial dissection of TORC1. In yeast cells, the vacuolar TORC1 stimulates protein synthesis and cell growth while the late endosomal TORC1 inhibits autophagy, an anti-aging process through which vacuoles (mammalian lysosomes) degrade damaged and obsolete proteins, lipids, and organelles for longevity [6]. This spatial dissection of TORC1 offers us a new route to explore how Osh6 contributes to longevity.
–
Osh6 transports phosphatidylinositol-4-phosphate (PI4P) from the plasma membrane (PM) to the endoplasmic reticulum (ER), where Sac1 dephosphorylates PI4P, and in turn transports phosphatidylserine (PS) from the ER to the PM [7]. Other Osh proteins also relay PI4P to Sac1 in the ER but extend the lifespan when depleted from the cell [8]. PS is a unique ligand of Osh6 and its closest paralog Osh7 [7]. Down-regulation of the PS synthesis in ER leads to fragmented vacuoles [9]. PS synthesized in the ER has to be transported to the Golgi and/or other organelles to control vacuolar morphology [10]. Disrupting the flip of PS from the lumen of the Golgi to the cytosolic side also leads to highly fragmented vacuoles [11]. A key enzyme for such flipping is Drs2 [12]. The fact that up-regulated Osh6 promotes vacuole fusion [5] and that Osh6 transports PS and PI4P [7] led us to hypothesize that Osh6 may mediate PS/PI4P transport in intracellular membranes.
–
In support of the above hypothesis, we found an accumulation of PI4P on the Golgi in cells of PERG6–OSH6. The accumulation site is likely the trans-Golgi network (TGN) since PERG6–OSH6 mimicked mutants defective in Golgi PS and PI4P trafficking [12] in rerouting a portion of the PM protein Pma1 to vacuoles. Intriguingly, this rerouting of Pma1 was dependent on the TGN-to-LE trafficking pathway. Based on these new findings and other published results, we propose that up-regulation of Osh6 speeds up vesicle trafficking between the TGN and LE by adjusting the local concentration of PI4P and PS on the TGN.
RESULTS
OSH6 genetically interacts with DRS2, whose protein works on the Golgi
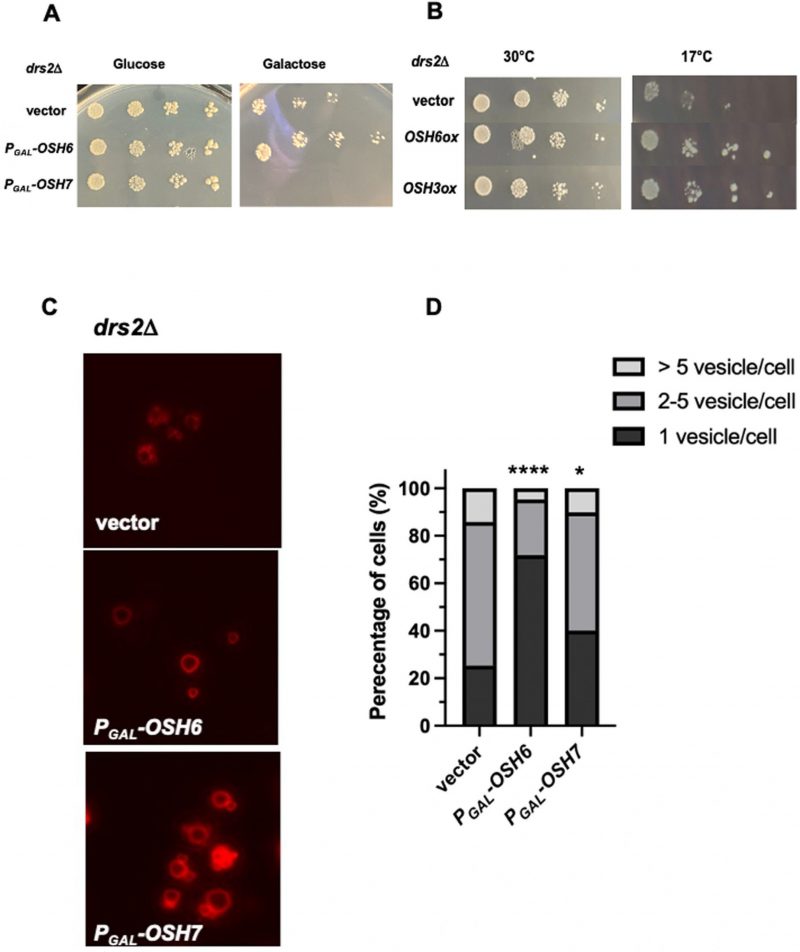
To explore how Osh6 affects vacuoles and longevity, we tested potential genetic interactions between OSH6 and genes involved in the metabolism of PS and PI4P, ligands of Osh6. Drs2 flips PS from the lumen of the Golgi to the cytosolic side and leads to fragmented vacuoles when mutated [11][12]. We over-expressed OSH6 and other OSH genes in a drs2Δ mutant (Fig. 1A, 1B). Interestingly, over-expression of OSH6 by a galactose promoter assisted the growth of drs2Δ cells marginally but over-expression of its closest paralog OSH7 severely delayed the cell growth at 30°C (Fig. 1A). At 17°C, over-expression of OSH6 or OSH3 by a high copy plasmid complemented the cold sensitivity of drs2Δ (Fig. 1B). We further checked the vacuolar morphology of the drs2Δ mutants with over-expressed OSH genes and found that over-expression of OSH6 significantly promoted vacuole fusion in drs2Δ while over-expression of OSH7 only marginally affected the morphology (Figs. 1C, 1D, and Fig. S1A).
–
–
Different from their effects on drs2Δ, over-expression of OSH6/7 did not obviously affect the growth of sac1Δ (Fig. S1B). For vacuolar morphology, over-expression of OSH6 did not show an obvious impact on sac1Δ (Fig. S1C). The different impact of OSH6 and OSH7 genes on drs2Δ and sac1Δ mutants suggest that Osh6 and Osh7 proteins have their own working locations in addition to their common ER-PM contact site. To search for such unique working places of Osh6, we then monitored the localization of PS and PI4P in cells of different mutants.
Up-regulation of OSH6 causes accumulation of PI4P on the Golgi
We first checked the localization of PS by the GFP-Lact-C2 probe, which was already used by many other labs [10][13]. To use this plasmid, we upregulated the expression of OSH7 by replacing its endogenous promoter on its chromosome by a short version of the promoter of ERG6 to obtain PERG6–OSH7 as we did for the up-regulation of OSH6 [5]. Similar to a previous report [13], up-regulation of Osh6 enriched PS on the PM of small buds (Fig. S2). We did not notice any obvious intracellular accumulations of PS in either PERG6–OSH6 or PERG6–OSH7 cells when compared with wild type cells (Fig. S2). This lack of differences in PS localization seems inconsistent with the genetic interactions between OSH6 and DRS2 (Fig. 1). This apparent inconsistency is likely due to the limitation of our current assay method for PS localization.
–
Then we monitored the localization of PI4P. We chose the probe 2XPH-OSBP-GFP because it tends to bind Golgi localized PI4P due to the pH of the cytoplasm [14][15][16]. Like previously reported, 2XPH-OSBP-GFP visualized PI4P-decorated punctates, typical Golgi structures (Fig. 2A). In the wild type, some cells showed plasma membrane PI4P with this probe (see arrowhead-pointed cell in Fig. 2A). Expectedly, deletion of Sac1 (sac1Δ) accumulated PI4P (Figs. 2A, 2B). Interestingly, PERG6–OSH6 accumulated PI4P inside cells (Figs. 2A, 2B). PERG6–OSH7 cells also showed punctate PI4P, but with less brightness (Fig. 2A, 2B). The fraction of cells carrying high intracellular PI4P levels was much higher in sac1Δ and PERG6–OSH6 than that in wild type and PERG6–OSH7 (Fig. 2B).
–
–
A Fisher Exact test showed that the fraction of cells with high PI4P intensity (>50 AU/cell) in sac1Δ (31/64) (p=0.043) and PERG6–OSH6 (42/83) (p=0.016) are significantly higher than that in wild type. Although both PERG6–OSH6 and sac1Δ accumulated PI4P, they distribute PI4P differentially. PERG6–OSH6 only showed punctate structures of PI4P, while sac1Δ cells also had PI4P on large membrane encircled organelles (arrow-pointed structure in Fig. 2A). Moreover, about 1/2 of sac1Δ cells showed bud-enriched PI4P, whereas only about 1/5 of PERG6–OSH6 cells showed such an enrichment (Fig. 2C). In wild type cells, only about 1/10 of cells had such bud-enriched PI4P. The polarized secretion to the budding tip and small buds depends on dephosphorylation of PI4P by Sac1 [17]. Deletion of Sac1 likely caused an accumulation of PI4P in polarized secretory vesicles. The observation that PERG6–OSH6 had a mild accumulation of PI4P in small buds (Fig. 2D) suggests that PERG6–OSH6 has more polarized secretion and/or partial defect in dephosphorylation of PI4P in small budded cells. In support, PERG6–OSH6 cells have more polarized actin cables [5], along which secretory vesicles are transported.
–
To check whether the punctate PI4P is on the Golgi in PERG6–OSH6, we integrated the Sec7-mCherry coding DNA into the URA3 gene [18] in PERG6–OSH6 and PERG6–OSH7 cells and then checked the colocalization of PI4P and Sec7 (a Golgi marker; Fig. 2D). In both PERG6–OSH6 and PERG6–OSH7 cells, PI4P did overlap with the Sec7-mCherry (see arrowhead pointed structures in Fig. 2D), suggesting that up-regulation of Osh6 enriches PI4P on the Golgi. The higher enrichment of PI4P on the Golgi in PERG6–OSH6 than in PERG6–OSH7 may be a contributor for longevity, since PERG6–OSH7 did not extend the replicative lifespan (data not shown).
Up-regulation of OSH6 delays the Pma1 secretion, a function of the Golgi
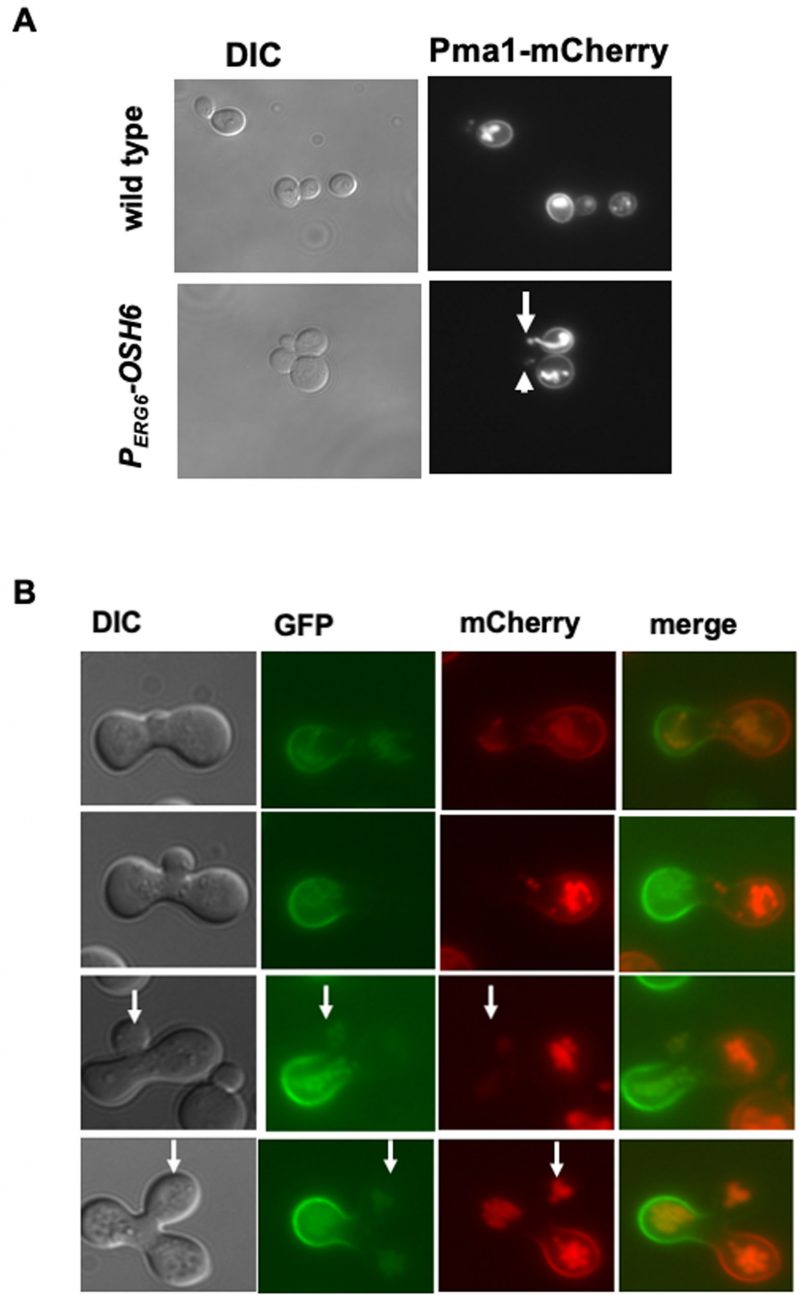
Since both PS and PI4P on the Golgi are required for proper secretion of the PM protein Sur7 and Pma1 [12], the accumulation of PI4P on the Golgi in PERG6–OSH6 (Fig. 2) may affect the secretion. Thus, we monitored the localization of PM proteins. For this purpose, we constructed PERG6–OSH6 Sur7-GFP and PERG6–OSH6 Pma1-mCherry strains by mating PERG6–OSH6 with Sur7-GFP or Pma1-mCherry strains [19]. Up-regulation of Osh6 (PERG6–OSH6) did not show obvious effects on the PM Sur7 (Fig. S3), but affected the secretion of Pma1 (Fig. 3A). First, PERG6–OSH6 cells accumulated more Pma1 inside cells; almost every cell had intracellular Pma1. The intracellular accumulation sites of Pma1 are likely in vacuoles, since the structure of the Pma1-accumulated organelle in some small-budded cells is very similar to vacuolar segregation structure during budding (see arrow pointed structure in Fig. 3A). Second, many small buds of PERG6–OSH6 cells did not show discernible Pma1 (see arrowhead-pointed buds in Fig. 3A), suggesting an alteration in secretion and/or endocytosis in the bud.
–
–
To double check the effect of up-regulation of Osh6 on Pma1 in small buds, we conducted a zygote assay (Fig. 3B). We mated wild type Pma1-GFP with PERG6–OSH6 Pma1-mCherry and then monitored the fluorescence of zygotes. As shown in Fig. 3B, the Pma1-GFP signal from the wild type parent arrived at the zygote bud earlier than the Pma1-mCherry signal from the PERG6–OSH6 parent (see arrow-pointed buds in Fig. 3B), indicating that up-regulation of Osh6 delays the arrival of Pma1 to the bud PM. Moreover, the intracellular Pma1 in PERG6–OSH6 moved to the other parent and the bud, phenocopying what vacuoles do in such zygote assays [20].
–
Decreasing Golgi PI4P abrogates Osh6’s effects on Pma1 distribution
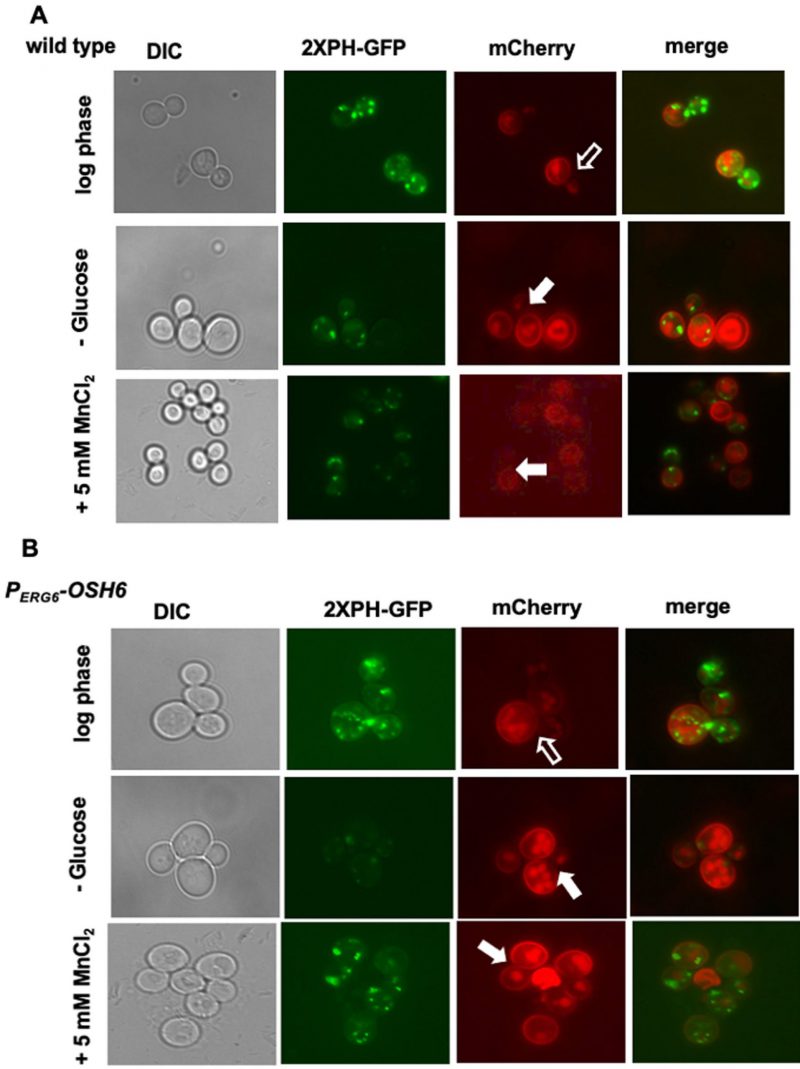
To confirm the Golgi localization of PI4P in PERG6–OSH6, we monitored the PI4P intensity after glucose starvation (Fig. 4). Glucose starvation reallocates the ER-localized Sac1 to the Golgi and prevents the accumulation of PI4P on the Golgi [21]. In line with this previous report, glucose starvation decreased the intensity of PI4P punctate in wild type cells in our assays (compare the GFP pictures of row 1 and row 2 in Fig. 4A). Upon glucose starvation, PERG6–OSH6 cells showed a clear decrease in PI4P intensity (compare the GFP pictures of row 1 and row 2 in Fig. 4B). In addition to Sac1 action, glucose starvation also changes cytosolic pH and decreases the binding of Golgi PI4P by PH domains [16]. Both possibilities (Sac1, pH) support that PI4P-decorated punctates in PERG6–OSH6 cells are Golgi structures.
–
–
Along with the decrease of the Golgi PI4P levels, the secretion pattern of Pma1 was also changed upon glucose starvation. In log phase cells grown in a medium with 2% glucose, the Pma1-mCherry signal on the PM of the mother-bud neck is not as bright as other areas of PM of the mother cell (see hollow arrow pointed areas in wild type (Fig. 4A) and PERG6–OSH6 (Fig. 4B)). Glucose starvation made Pma1 homogenously distributed on the mother PM (see solid arrow pointed cells in Fig. 4). Moreover, we found that treating yeast cells with 5 mM MnCl2 also decreased the Golgi PI4P level (Fig. 4) and simultaneously altered the secretion of Pma1 similar to what glucose starvation did (Fig. 4). These observations further confirm that up-regulation of Osh6 causes accumulation of PI4P on the Golgi.
The effects of up-regulation of OSH6 rely on the Golgi-to-LE trafficking
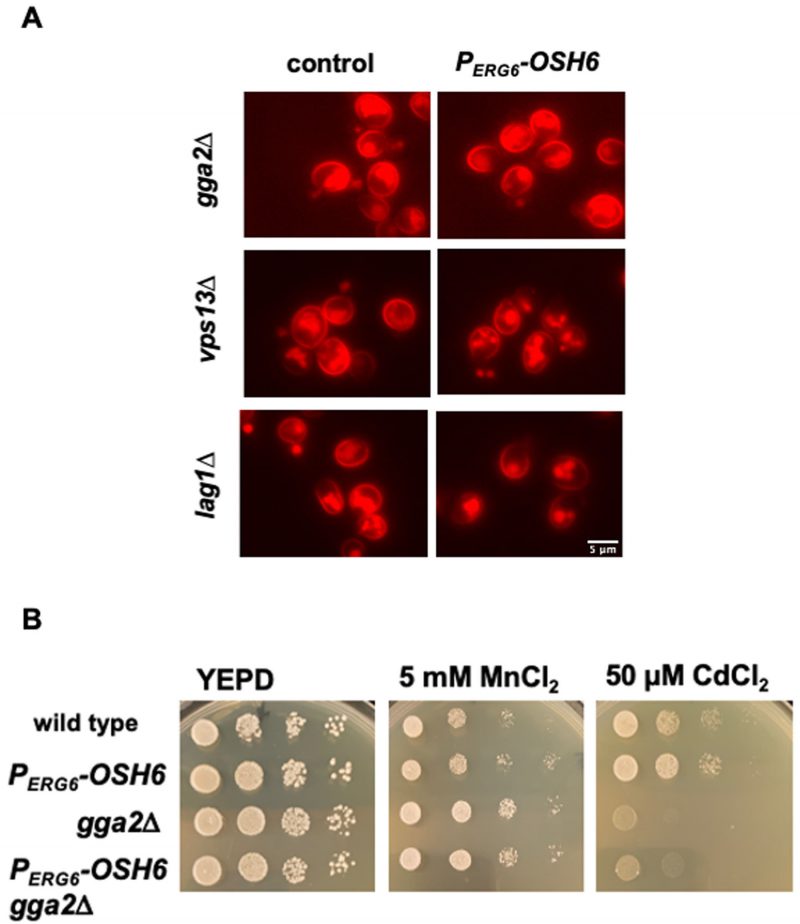
While multiple pathways transport post-Golgi vesicles to vacuoles, the pathway that depends on the level of Golgi PI4P is the TGN to LE (TGN-LE) pathway (also termed the CPY pathway) [22]. To test whether the TGN-LE pathway is used by Osh6 in rerouting Pma1 to vacuoles, we checked the impact of Gga2, which binds Golgi PI4P and catalyzes the formation of the CPY vesicles [22], and Vps13, which recycles materials from LE back to the Golgi [23] on the Pma1 localization in PERG6–OSH6 cells (Fig. 5). As a control, we also tested the impact of Lag1, a ceramide synthase working in the ER [24]. In the double mutant PERG6–OSH6 gga2Δ or PERG6–OSH6 vps13Δ, the Pma1 localization was similar to that in the corresponding gga2Δ or vps13Δ single mutant, where the intracellular Pma1 is dispersed to almost the whole cytoplasm (Fig. 5A). Similar to the trafficking of Pma1, a growth phenotype of PERG6–OSH6 gga2Δ also followed the pattern of the gga2Δ single mutant; PERG6–OSH6 gga2Δ was as resistant as gga2Δ to 5 mM MnCl2 and as sensitive as gga2Δ to 50 µM CdCl2 (Fig. 5B). The dependency of Osh6 on Gga2 supports that Osh6 works through the TGN-LE pathway. Different from gga2Δ or vps13Δ mutants, the PERG6–OSH6 lag1Δ double mutant behaved similar to PERG6–OSH6 in enriching Pma1 into certain organelles (Fig. 5A).
–
Up-regulation of OSH6 has a unique role in restoring cell growth
Pma1 is secreted to the PM by complexing with sphingolipids carrying very long chain fatty acids (VLCFA) in the ER and the Golgi [25]. Since sphingolipids with VLCFA are also required for functions of vacuolar ATPase and vacuolar morphology [26][27], we studied interactions between Osh6 and Sur4, which synthesizes VLCFA and controls vacuolar morphology via the TGN-LE trafficking step [28], and Lag1, which incorporates VLCFA into ceramide, the precursor for sphingolipids. Over-expressing OSH6 by a high copy plasmid restored vacuole fusion in lag1Δ and sur4Δ while over-expression of OSH4 did not show such restoration (Fig. 6A and Fig. S4). The observations that high levels of Osh6 down-regulated the secretion of Pma1 (Fig. 3) while promoted vacuole fusion in mutants (lag1Δ and sur4Δ defective in enzymes in the ER (Figs. 6A, 6B, and Fig. S4) further support the idea that Osh6 works on the Golgi.
–
–
To further test the genetic interactions between OSH6 and LAG1, we tested growth phenotype of the PERG6–OSH6 lag1Δ double mutant on caffeine plates (Fig. 6C). Caffeine inhibits TORC1 in a manner similar to rapamycin [29]. Shocking cells by 0.2% caffeine at 37°C for two days led to growth arrest (see middle panel of Fig. 6C). The growth was recovered after transferring this plate to room temperature (24°C) and incubating for five days (see row 1 and row 2 in Fig. 6C). Although lag1Δ showed a weak recovery, the PERG6-OSH6 lag1Δ double mutant recovered 10- to 100-fold better than lag1Δ did (rows 3 and 4 in Fig. 6C). Different from its effects on lag1Δ, PERG6–OSH6 only very mildly recovered the growth of a mutant missing Vps13, which facilitates the TGN-LE trafficking (rows 5 and 6 in Fig. 6C). The recovery of growth after TORC1 repression requires TORC1 activation to stimulate protein synthesis as well as TORC1 repression to relieve the inhibition on microautophagy, which affects vacuolar morphology [30]. Since LE TORC1 inhibits autophagy including microautophagy and vacuolar TORC1 stimulates cell growth [6], we interpret the recovery of growth of the PERG6–OSH6 lag1Δ mutant (Fig. 6C) and the promotion of vacuole fusion of Osh6 in lag1Δ and sur4Δ (Figs. 6A, 6B, and Fig. S4) as that up-regulation of Osh6 differentially affects the endosomal TORC1 and vacuolar TORC1 by altering the TGN-LE trafficking step.
DISCUSSION
While Osh6 and its homologs work on multiple organelles including the ER, the PM, secretory vesicles, and mitochondria [7][13][31][32], it differs from other Osh proteins in its effects on longevity. OSH6 extends the replicative lifespan when up-regulated, but other OSH genes (OSH1, OSH2, OSH3, OSH4) extend the lifespan when deleted from the genome [5][8]. The unique roles of Osh6 and the critical contribution of the TGN-LE trafficking to pro-longevity pathways [6] led us to propose that Osh6 accelerates the TGN-LE vesicle trafficking and differentially affects endosomal TORC1 and vacuolar TORC1 to increase longevity.
–
The TGN is a working place of Osh6
Our phenotypic and subcellular localization analyses along with other group’s results on Osh6 and PI4P suggest that a subset of Osh6 works on the TGN. Although both PERG6–OSH6 and sac1Δ accumulated PI4P inside cells, PERG6–OSH6 caused an enrichment of PI4P on Golgi-like punctate while in sac1Δ cells PI4P could also be found on other membranes that gave a faint signal (see arrow-pointed cell in Fig. 2A). In log phase cells, intracellular PI4P localizes to the Golgi membrane due to the function of the PI 4 kinase Pik1[33]. In sac1Δ cells, PI4P also localizes to other organelles including endosomal/vacuolar membranes and leads to a large vacuole with multiple invaginations [34] (also see Fig. S1C). Different from sac1Δ cells, PERG6–OSH6 cells have normal vacuolar morphology, two to five vacuolar vesicles per cell for most cells [5]. Thus, PERG6–OSH6 could not accumulate PI4P on endosomal/vacuolar membranes. Indeed, PI4P in PERG6–OSH6 overlapped with the Golgi marker Sec7 (Fig. 2D). The localization data of Osh6 from Orin’s lab show that Osh6 can work on TGN. A chromosomal version of GFP-tagged Osh6 with its endogenous promoter localizes to the PM and intracellular organelles in small buds [35]. During yeast cell division, organelles such as ER, late Golgi elements, and vacuoles are transported to buds at an early stage of budding [20][36][37]. Since ER hosts Sac1 and degrades PI4P, we conclude that the enriched PI4P in small buds of PERG6–OSH6 (Fig. 2C) is a sign of Osh6 working on late Golgi elements. In line with this conclusion, in PERG6–OSH6 cells a portion of Pma1 was rerouted to vacuoles (Fig. 3), phenocopying mutants missing Drs2 or Osh4, two proteins working on the TGN [12].
–
Osh6 promotes TGN-LE transport to extend the lifespan
Three lines of evidence suggest that elevated Osh6 promotes TGN-LE membrane trafficking as depicted in Fig. 7. First, PERG6–OSH6 caused an enrichment of PI4P on the Golgi (Fig. 2) and this relied on the TGN-LE protein Gga2, which binds Golgi PI4P for its effects (Fig. 5).
–
Second, over-expression of OSH6 restores vacuole fusion of mutants defective in TGN-LE trafficking. Deletion of Sur4 compromises the TGN-LE trafficking but does not affect the TGN-vacuole pathway [28]. In this work, we observed that over-expression of OSH6 but not OSH4 restored vacuole fusion in sur4Δ (Fig. S3) and lag1Δ (Fig. 4), cells which are defective in incorporating Sur4’s product into sphingolipids. The specific growth recovery effect of PERG6–OSH6 on lag1Δ (Fig. 6) suggests that the TGN-LE trafficking step is the most likely route for Osh6 to control vacuolar morphology and TORC1.
–
Third, the effect of up-regulation of Osh6 on Pma1 localization is dependent on Gga2 and Vps13, key players in the TGN-LE transport (Fig. 5). Moreover, VPS13 and OSH6 share two genetic interaction partners, CSG2 and VPS35, and GGA2 also interacts with CSG2 [38][39]. Csg2 is required for the synthesis of mannosyl-inositol-phosphorylceramide, a complex sphingolipid. Vps35 is required for the recycling of vesicles from endosomes back to the TGN and genetically interacts with Tor1 [40]. Thus, a simple model reconciling all phenotypes of PERG6–OSH6 is that elevated Osh6 accelerates the TGN-LE transport and in turn LE-vacuole transport (Fig. 7B). Such acceleration limits the membrane size of LE so that LE can only host minimal endosomal TORC1.
–
 | FIGURE 7: A working model for Osh6's role in TGN-LE trafficking. (A) A summary of the maturation of the trans-Golgi network (TGN) and post-Golgi trafficking. The formation of Pma1-vesicles for secretion occurs at late cisternae of the TGN while vesicles carrying carboxylpeptidase Y (CPY) or other vacuolar hydrolases to late endosome (LE) occurs in early cisternae of the TGN [41] (see text for detail). (B) A hypothetical action of up-regulated Osh6. Up-regulation of Osh6 transports more PI4P to the early cisternae of the TGN, which facilitates the TGN-to-LE trafficking since the formation of the CPY-carrying vesicles depends on Gga2-PI4P binding. The accelerated TGN-LE trafficking refurbishes fresh lipids to LE and vacuoles and thus support vacuolar functions. As a side effect, transporting PI4P out of the late cisternae of TGN delays the secretion of Pma1 and leads to the rerouting of Pma1 to LE and vacuoles. |
–
Osh6 may accelerate TGN-LE transport by adjusting the local PS and PI4P concentrations of the early TGN
Osh6’s two lipid ligands, PS and PI4P, are both crucial for the formation of post-Golgi vesicles and post-Golgi membrane trafficking. Upon maturation of the trans-Golgi cisternae, vesicles carrying Pma1 and other cargoes are budded from the late TGN cisternae and transported to the PM (summarized in Fig. 7A). The Pma1-vesicles require balanced PI4P and PS [12]. During the process of TGN maturation, the early cisternae of the TGN forms vesicles destined to LE [41]. The formation of TGN-LE vesicles depends on Gga2-PI4P interactions [22]. In active growing cells, the TGN-LE transport is very slow [42]. This slow rate is likely caused by lack of PI4P on the early TGN cisternae, since the majority of Golgi PI4P is used for the formation of secretory vesicles and is consumed by Osh4 and other Osh proteins during secretion [12][17][31].
–
Elevated Osh6 could accelerate the TGN-LE transport by its PI4P/PS swapping activity (Fig. 7B). The formation of vesicles toward LE requires PI4P [22]. Moreover, PS on LE needs to be transported back to the TGN by the LE-TGN retrograde pathway [43][44], since elevated PS or PS/phosphatidylethanolamine (PE) ratio on LE and vacuolar membranes leads to fragmented vacuoles [45]. Osh6’s working on TGN may transport PI4P to the early TGN cisternae and remove PS from that area (dark arrows in Fig. 7B). A support for such removal of PS is the observation that over-expression of OSH6 complemented defects of drs2Δ (Fig. 1). This complementation suggests that Osh6 can glean PS for some essential Golgi functions that are normally achieved by Drs2-flipped PS. The potential PI4P/PS swapping on the TGN facilitates both the formation of vesicles toward LE and the LE-to-TGN retrograde trafficking. Such elevated retrograde trafficking also leads to a decrease of LE membrane and thus endosomal TORC1. Testing this model in future studies would reveal detailed mechanistic links between TGN-LE trafficking and endosomal/vacuolar TORC1 activities and hence provide insights on how oxysterol-binding proteins control TORC1 and longevity in other organisms.
MATERIALS AND METHODS
Strains, plasmids, yeast manipulations, and media
All yeast strains (listed in Table S1) are derivatives of BY4742. The PS-labeling GFP-Lact-C2 plasmid was kindly provided by Drs. Gregory Fairn and Vanina Zaremberg. The PI4P-labeling 2XPH-OSBP-GFP plasmid pPGK1303 was kindly provided by Dr. Christina Mitchell. OSH plasmids (pCB237 (Yep24-OSH6), pCB238 (Yep24-OSH3), pCB241 (Yep24-OSH4), pCB247 (PGAL-OSH7), pCB248 (PGAL-OSH6)) were kindly provided by Dr. Christopher Beh. Construction of PERG6–OSH7 is described below. Double and triple mutants of PERG6–OSH6 carrying Pma1-mCherry or Sur7-GFP were constructed by standard yeast mating of PERG6–OSH6 with Pma1-mCherry, Sur7-GFP strains provided by Dr. Mara Duncan [19], induction of meiosis, and tetrad dissection. Standard yeast media YEPD, yeast extract (1%)-peptone (2%)-dextrose (2%), was used unless otherwise stated.
Construction of PERG6-OSH7 mutants
The strategies for constructing previously described PERG6–OSH6 mutants [5] were employed to construct the PERG6–OSH7 in this study. The PERG6–OSH7 strain was constructed by switching the endogenous promoter of OSH7 with the promoter of ERG6 via in vivo recombination. The promoter of ERG6 on the plasmid pRS316-ERG6 [46] was PCR-amplified by the primer Perg6OSH7up (5′TATCAGTATATTATAGGAATGTTAATTCGCTCGTGCGTACTAATTTTTGATTCGGTAATCTCC) and Perg6OSH7 down (5′ACTGTTTGTTAAAGAAGGTATATTCTTTAGTTTATTGAGAGCCATCATCTTATGCTGCCTACT). This PCR-amplified fragment contained the URA3 gene followed by the basal ERG6 promoter flanked by sequences homologous to the OSH7 promoter. This fragment was transformed into BY4742. Verification of the correct insertion-replacement was confirmed by PCR with primers RCERG6up (5’ATAGTTCGGGTGTTTT) and RCosh7down (5’TGATCTGTTCTTCATG). A 1.5 kb PCR-amplified fragment was used as the diagnostic band for the PERG6–OSH7 replacement.
–
PS, PI4P, and vacuole labeling
A plasmid expressing the GFP-Lact-C2 fusion protein, which binds PS [10] or a plasmid expressing PI4P-binding marker proteins (pPGK1303/PH-OSBP-GFP) [15] was transformed into the wild type BY4742 or different mutant strains. Transformation was done with the LiAc protocol following all the steps listed in [47]. If the mutant strain carried the URA3-PERG6 promoter, the URA3 marker was replaced by a Kanamycin-resistance marker with BamHI-linearized M3927 obtained from Addgene [48] before PS and PI4P assay. Transformants were grown in SC-URA liquid media at 30°C to early log phase (OD600 between 0.2 to 0.4). Cells were observed under a 90i eclipse microscope and photographed under the FITC filter for GFP and Texas Red filter for mCherry. The resulting pictures were imported to the ImageJ software to report the fluorescent intensity of the region of interest (bud, mother) that was drawn manually. Intensities obtained for each cell were used for distribution analyses.
–
Vacuoles were labeled by FM4-64 and chased as described before [49].
–
The co-localization of PI4P with the Golgi marker Sec7-mCherry
The URA3::Sec7-mCherry DNA linearized by XcmI and BstB1 from YIplac211-SEC7-mCherry2Bx [18] was transformed into URA3-PERG6–OSH6 (FTY373) and URA3-PERG6–OSH7 (FTY437) cells. The resulting transformants were selected on SC+URA+5-fluroorotic acid plate. After confirming the lack of growth on SC-URA plates, the Sec7-mCherry version of PERG6–OSH6 (FTY624) and PERG6–OSH7 (FTY625) strains were transformed by the plasmid carrying the PI4P-binding probe (2XPH-OSBP-GFP). The resulting transformants were grown to early log phase in SC-URA and then photographed under the FITC (for PI4P) and Texas Red (for Sec7) filters.
–
Image quantitation and statistical analyses
All the pictures were analyzed by the ImageJ software to measure the fluorescent intensity of the region of interest (bud, mother) that was drawn. For quantitative analysis, we used Graph Pad Prism 8 software to generate all the graphs in this study. The significance of the difference in the mean values was determined by 1 WAY ANOVA that was applied on all the data by using Tukey’s multiple comparisons test, all those tests indicated P value < 0.0001 was considered as significant. Also, other statistics analysis such as Fisher exact test was conducted to judge whether the fraction of cells with bud-enriched PI4P is significant between two samples.
REFERENCES
- Pietrangelo A and Ridgway ND (2018). Bridging the molecular and biological functions of the oxysterol-binding protein family. Cell Mol Life Sci 75(17):3079-3098. 10.1007/s00018-018-2795-y
- Beh CT, Rine J (2004). A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J Cell Sci 117(Pt 14):2983-96. 10.1242/jcs.01157
- Kobuna H, Inoue T, Shibata M, Gengyo-Ando K, Yamamoto A, Mitani S, Arai H (2010). Multivesicular body formation requires OSBP-related proteins and cholesterol. PLoS Genet 6(8):e1001055. 10.1371/journal.pgen.1001055
- Birnbaum A, Sodders M, Bouska M, Chang K, Kang P, McNeill E, Bai H (2021). FOXO Regulates Neuromuscular Junction Homeostasis During Drosophila Aging. Front Aging Neurosci 12:567861. 10.3389/fnagi.2020.567861
- Gebre S, Connor R, Xia Y, Jawed S, Bush JM, Bard M, Elsalloukh H, Tang F (2012). Osh6 overexpression extends the lifespan of yeast by increasing vacuole fusion. Cell Cycle 11(11):2176-88. 10.4161/cc.20691
- Hatakeyama R, Péli-Gulli MP, Hu Z, Jaquenoud M, Garcia Osuna GM, Sardu A, Dengjel J, De Virgilio C (2019). Spatially Distinct Pools of TORC1 Balance Protein Homeostasis. Mol Cell 73(2):325-338.e8. 10.1016/j.molcel.2018.10.040
- Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC (2013). Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature 501(7466):257-61. 10.1038/nature12430
- Huang J, Mousley CJ, Dacquay L, Maitra N, Drin G, He C, Ridgway ND, Tripathi A, Kennedy M, Kennedy BK, Liu W, Baetz K, Polymenis M, Bankaitis VA (2018). A Lipid Transfer Protein Signaling Axis Exerts Dual Control of Cell-Cycle and Membrane Trafficking Systems. Dev Cell 44(3):378-391.e5. 10.1016/j.devcel.2017.12.026
- Kannan M, Lahiri S, Liu LK, Choudhary V, Prinz WA (2017). Phosphatidylserine synthesis at membrane contact sites promotes its transport out of the ER. J Lipid Res 58(3):553-562. 10.1194/jlr.M072959
- Ganesan S, Sosa Ponce ML, Tavassoli M, Shabits BN, Mahadeo M, Prenner EJ, Terebiznik MR, Zaremberg V (2019). Metabolic control of cytosolic-facing pools of diacylglycerol in budding yeast. Traffic 20(3):226-245. 10.1111/tra.12632
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K (2007). Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol Biol Cell 18(1):295-312. 10.1091/mbc.e06-05-0461
- Hankins HM, Sere YY, Diab NS, Menon AK, Graham TR (2015). Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol Biol Cell 26(25):4674-85. 10.1091/mbc.E15-07-0487
- D’Ambrosio JM, Albanèse V, Čopič A (2019). Following Anterograde Transport of Phosphatidylserine in Yeast in Real Time. Methods Mol Biol 1949:35-46. 10.1007/978-1-4939-9136-5_4
- Levine TP, Munro S (2002). Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol 12(9):695-704. 10.1016/s0960-9822(02)00779-0
- Wiradjaja F, Ooms LM, Tahirovic S, Kuhne E, Devenish RJ, Munn AL, Piper RC, Mayinger P, Mitchell CA (2007). Inactivation of the phosphoinositide phosphatases Sac1p and Inp54p leads to accumulation of phosphatidylinositol 4,5-bisphosphate on vacuole membranes and vacuolar fusion defects. J Biol Chem 282(22):16295-307. 10.1074/jbc.M701038200
- Shin JJH, Liu P, Chan LJ, Ullah A, Pan J, Borchers CH, Burke JE, Stefan C, Smits GJ, Loewen CJR (2020). pH Biosensing by PI4P Regulates Cargo Sorting at the TGN. Dev Cell 52(4):461-476.e4. 10.1016/j.devcel.2019.12.010
- Ling Y, Hayano S, Novick P (2014). Osh4p is needed to reduce the level of phosphatidylinositol-4-phosphate on secretory vesicles as they mature. Mol Biol Cell 25(21):3389-400. 10.1091/mbc.E14-06-1087
- Day KJ, Casler JC, Glick BS (2018). Budding Yeast Has a Minimal Endomembrane System. Dev Cell 44(1):56-72.e4. 10.1016/j.devcel.2017.12.014
- Lang MJ, Martinez-Marquez JY, Prosser DC, Ganser LR, Buelto D, Wendland B, Duncan MC (2014). Glucose starvation inhibits autophagy via vacuolar hydrolysis and induces plasma membrane internalization by down-regulating recycling. J Biol Chem 289(24):16736-47. 10.1074/jbc.M113.525782
- Weisman LS, Wickner W (1988). Intervacuole exchange in the yeast zygote: a new pathway in organelle communication. Science 241(4865):589-91. 10.1126/science.3041591
- Piao H, MacLean Freed J, Mayinger P (2012). Metabolic activation of the HOG MAP kinase pathway by Snf1/AMPK regulates lipid signaling at the Golgi. Traffic 13(11):1522-31. 10.1111/j.1600-0854.2012.01406.x
- Demmel L, Gravert M, Ercan E, Habermann B, Müller-Reichert T, Kukhtina V, Haucke V, Baust T, Sohrmann M, Kalaidzidis Y, Klose C, Beck M, Peter M, Walch-Solimena C (2008). The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit. Mol Biol Cell 19(5):1991-2002. 10.1091/mbc.e06-10-0937
- De M, Oleskie AN, Ayyash M, Dutta S, Mancour L, Abazeed ME, Brace EJ, Skiniotis G, Fuller RS (2017). The Vps13p-Cdc31p complex is directly required for TGN late endosome transport and TGN homotypic fusion. J Cell Biol 216(2):425-439. 10.1083/jcb.201606078
- Megyeri M, Prasad R, Volpert G, Sliwa-Gonzalez A, Haribowo AG, Aguilera-Romero A, Riezman H, Barral Y, Futerman AH, Schuldiner M (2019). Yeast ceramide synthases, Lag1 and Lac1, have distinct substrate specificity. J Cell Sci 132(12):jcs228411. 10.1242/jcs.228411
- Gaigg B, Timischl B, Corbino L, Schneiter R (2005). Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J Biol Chem 280(23):22515-22. 10.1074/jbc.M413472200
- Chung JH, Lester RL, Dickson RC (2003). Sphingolipid requirement for generation of a functional v1 component of the vacuolar ATPase. J Biol Chem 278(31):28872-81. 10.1074/jbc.M300943200
- Seeley ES, Kato M, Margolis N, Wickner W, Eitzen G (2002). Genomic analysis of homotypic vacuole fusion. Mol Biol Cell 13(3):782-94. 10.1091/mbc.01-10-0512
- Obara K, Kojima R, Kihara A (2013). Effects on vesicular transport pathways at the late endosome in cells with limited very long-chain fatty acids. J Lipid Res 54(3):831-42. 10.1194/jlr.M034678
- Reinke A, Chen JC, Aronova S, Powers T (2006). Caffeine targets TOR complex I and provides evidence for a regulatory link between the FRB and kinase domains of Tor1p. J Biol Chem 281(42):31616-26. 10.1007/s00018-018-2795-y
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C (2005). The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell 19(1):15-26. 10.1016/j.molcel.2005.05.020
- Smindak RJ, Heckle LA, Chittari SS, Hand MA, Hyatt DM, Mantus GE, Sanfelippo WA, Kozminski KG (2017). Lipid-dependent regulation of exocytosis in S. cerevisiae by OSBP homolog (Osh) 4. J Cell Sci 130(22):3891-3906. 10.1242/jcs.205435
- Tian S, Ohta A, Horiuchi H, Fukuda R (2018). Oxysterol-binding protein homologs mediate sterol transport from the endoplasmic reticulum to mitochondria in yeast. J Biol Chem 293(15):5636-5648. 10.1074/jbc.RA117.000596
- Faulhammer F, Kanjilal-Kolar S, Knödler A, Lo J, Lee Y, Konrad G, Mayinger P (2007). Growth control of Golgi phosphoinositides by reciprocal localization of sac1 lipid phosphatase and pik1 4-kinase. Traffic 8(11):1554-67. 10.1111/j.1600-0854.2007.00632.x
- Foti M, Audhya A, Emr SD (2001). Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell 12(8):2396-411. 10.1091/mbc.12.8.2396
- Lipp NF, Gautier R, Magdeleine M, Renard M, Albanèse V, Čopič A, Drin G (2019). An electrostatic switching mechanism to control the lipid transfer activity of Osh6p. Nat Commun 10(1):3926. 10.1038/s41467-019-11780-y
- Rossanese OW, Reinke CA, Bevis BJ, Hammond AT, Sears IB, O’Connor J, Glick BS (2001). A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J Cell Biol 153(1):47-62. 10.1083/jcb.153.1.47
- Arai S, Noda Y, Kainuma S, Wada I, Yoda K (2008). Ypt11 functions in bud-directed transport of the Golgi by linking Myo2 to the coatomer subunit Ret2. Curr Biol 18(13):987-91. 10.1016/j.cub.2008.06.028
- Costanzo M, et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science 353(6306):aaf1420. 10.1126/science.aaf1420
- Surma MA, Klose C, Peng D, Shales M, Mrejen C, Stefanko A, Braberg H, Gordon DE, Vorkel D, Ejsing CS, Farese R Jr, Simons K, Krogan NJ, Ernst R (2013). A lipid E-MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell 51(4):519-30. 10.1016/j.molcel.2013.06.014
- Aronova S, Wedaman K, Anderson S, Yates J 3rd, Powers T (2007). Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae. Mol Biol Cell 18(8):2779-94. 10.1091/mbc.e07-03-0274
- Casler JC, Glick BS (2020). A microscopy-based kinetic analysis of yeast vacuolar protein sorting. Elife 9:e56844. 10.7554/eLife.56844
- Ha SA, Torabinejad J, DeWald DB, Wenk MR, Lucast L, De Camilli P, Newitt RA, Aebersold R, Nothwehr SF (2003). The synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol Biol Cell 14(4):1319-33. 10.1091/mbc.e02-10-0686
- Ma M, Kumar S, Purushothaman L, Babst M, Ungermann C, Chi RJ, Burd CG (2018). Lipid trafficking by yeast Snx4 family SNX-BAR proteins promotes autophagy and vacuole membrane fusion. Mol Biol Cell 29(18):2190-2200. 10.1091/mbc.E17-12-0743
- Tani M, Kuge O (2012). Involvement of complex sphingolipids and phosphatidylserine in endosomal trafficking in yeast Saccharomyces cerevisiae. Mol Microbiol 86(5):1262-80. 10.1111/mmi.12057
- Wu Y, Takar M, Cuentas-Condori AA, Graham TR (2016). Neo1 and phosphatidylethanolamine contribute to vacuole membrane fusion in Saccharomyces cerevisiae. Cell Logist 6(3):e1228791. 10.1080/21592799.2016.1228791
- Tedrick K, Trischuk T, Lehner R, Eitzen G (2004). Enhanced membrane fusion in sterol-enriched vacuoles bypasses the Vrp1p requirement. Mol Biol Cell 15(10):4609-21. 10.1091/mbc.e04-03-0194
- Reece-Hoyes JS, Walhout AJM (2018). High-Efficiency Yeast Transformation. Cold Spring Harb Protoc 2018(7). 10.1101/pdb.prot094995
- Voth WP, Jiang YW, Stillman DJ (2003). New ‘marker swap’ plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20(11):985-93. 10.1002/yea.1018
- Tang F, Kauffman EJ, Novak JL, Nau JJ, Catlett NL, Weisman LS (2003). Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature 422(6927):87-92. 10.1038/nature01453
–
SUPPLEMENTAL INFORMATION
![]() Download Supplemental Information
Download Supplemental Information
ACKNOWLEDGMENTS
We thank Drs. Gregory Fairn and Va-nina Zaremberg for the PS labeling GFP-Lact-C2 plasmid, Dr. Chrstina Mitchell for the PI4P-labeling 2XPH-OSBP-GFP plasmid, Dr. Christopher Beh for OSH plasmids, Dr. Mara Duncan for yeast strains DLY046 and QAY559, and Dr. Todd Graham for the yeast strain HHY103. We thank Drs. Suriakarthiga Ganesan and Vanina Zaremberg for discussion on Pma1 localization and Dr. Benjamin Glick for instructions on integrating the Sec7-mCherry into the URA3 locus. This project was partly supported by the office of research and sponsored program (ORSP) of University of Arkansas at Little Rock ORSP Seed grant "Osh6 – mediated sterol redistribution extends lifespan” (to FT) and by the ORSP Signature Experience Grant “Osh6 enhances the CPY pathway to extend the lifespan” (to IK).
Microscopes used in this study was partly supported by the Arkansas INBRE program, supported by a grant from the National Institute of Gen-eral Medical Sciences, (NIGMS), P20 GM103429 from the National Institutes of Health.”
COPYRIGHT
© 2022

Up-regulation of Osh6 boosts an anti-aging membrane trafficking pathway toward vacuoles by Kadhim et al. is licensed under a Creative Commons Attribution 4.0 International License.