
Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.

Luminal acetylation of microtubules is not essential for Plasmodium berghei and Toxoplasma gondii survival
Acetylation of α-tubulin at lysine 40 is not essential for cytoskeletal stability in Plasmodium berghei or Toxoplasma gondii, suggesting redundancy and plasticity in microtubule regulation in these parasites.

The dual-site agonist for human M2 muscarinic receptors Iper-8-naphtalimide induces mitochondrial dysfunction in Saccharomyces cerevisiae
S. cerevisiae is a model to study human GPCRs. N-8-Iper, active against glioblastoma via M2 receptor, causes mitochondrial damage in yeast by binding Ste2, highlighting evolutionary conservation of GPCRs.

Integrative Omics reveals changes in the cellular landscape of peroxisome-deficient pex3 yeast cells
To uncover the consequences of peroxisome deficiency, we compared Saccharomyces cerevisiae wild-type with pex3 cells, which lack peroxisomes, employing quantitative proteomics and transcriptomics technologies.

Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
Rebekkah E. Pope1, Patrick Ballmann2, Lisa Whitworth3 and Rolf A. Prade1,*
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
Evelyn Tevere1,a, María G. Mediavilla1,a, Cecilia B. Di Capua1, Marcelo L. Merli1, Carlos Robello2,3, Luisa Berná2,4 and Julia A. Cricco
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.
Sir2 regulates selective autophagy in stationary-phase yeast cells
Ji-In Ryua, Juhye Junga, and Jeong-Yoon Kim
This study establishes Sir2 as a previously unrecognized regulator of selective autophagy during the stationary phase and highlight how cells dynamically control organelle degradation.
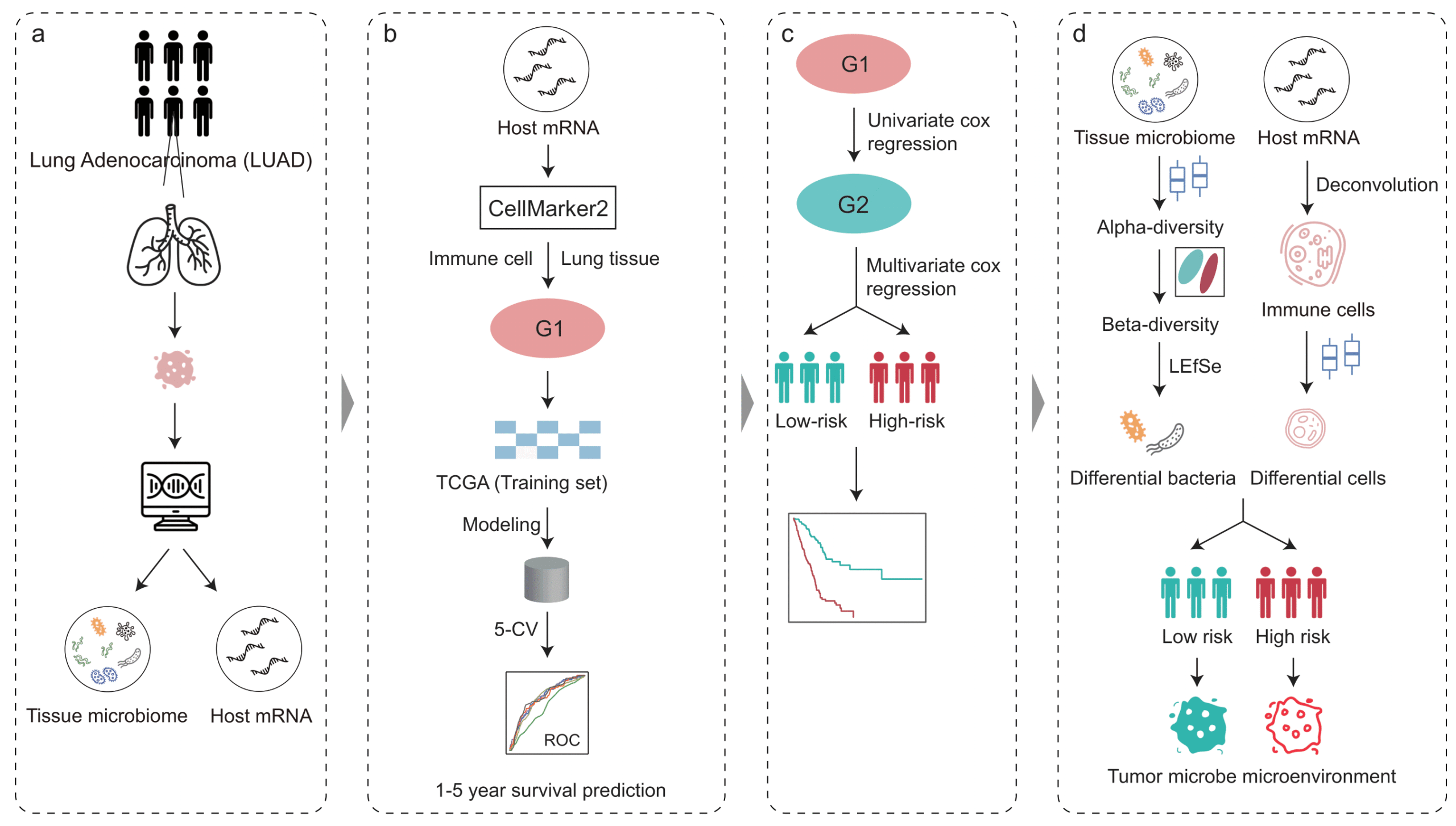
Predictable regulation of survival by intratumoral microbe-immune crosstalk in patients with lung adenocarcinoma
Shuo Shi1, Yuwen Chu2,3, Haiyan Liu4,5, Lan Yu6,7,8, Dejun Sun8,9, Jialiang Yang2,3,5, Geng Tian2,3, Lei Ji2,3, Cong Zhang10 and Xinxin Lu11
Intratumoral microbiota can regulate the tumor immune microenvironment (TIME) and mediate tumor prognosis by promoting inflammatory response or inhibiting anti-tumor effects. Our study demonstrated that intratumoral microbiota-immune crosstalk was strongly associated with prognosis in LUAD patients, which would provide new targets for the development of precise therapeutic strategies.

The last two transmembrane helices in the APC-type FurE transporter act as an intramolecular chaperone essential for concentrative ER-exit
Yiannis Pyrris1, Georgia F. Papadaki1, Emmanuel Mikros2 and George Diallinas1,3
FurE is a H+ symporter specific for the cellular uptake of uric acid, allantoin, uracil, and toxic nucleobase analogues in the fungus Aspergillus nidulans. Being member of the NCS1 protein family, FurE is structurally related to the APC-superfamily of transporters.

Basal level of ppGpp coordinates Escherichia coli cell heterogeneity and ampicillin resistance and persistence
Paulina Katarzyna Grucela1 and Yong Everett Zhang1
The universal stringent response alarmone ppGpp (guanosine penta and tetra phosphates) plays a crucial role in various aspects of fundamental cell physiology (e.g., cell growth rate, cell size) and thus bacterial tolerance to and survival of external stresses, including antibiotics. In tihs study, we discuss the fundamental role of basal level of ppGpp in regulating cell homogeneity and ampicillin persistence.
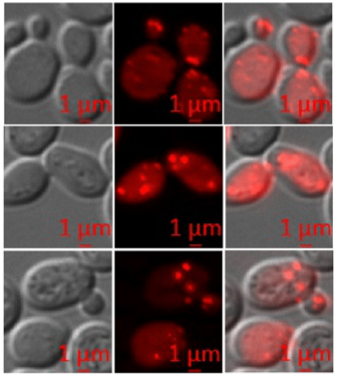
Investigation of the acetic acid stress response in Saccharomyces cerevisiae with mutated H3 residues
Nitu Saha1, Swati Swagatika1 and Raghuvir Singh Tomar1
Yeast cells respond to acetic acid in diverse ways. Here, we have elucidated the deleterious effects of acetic acid on different histone mutants
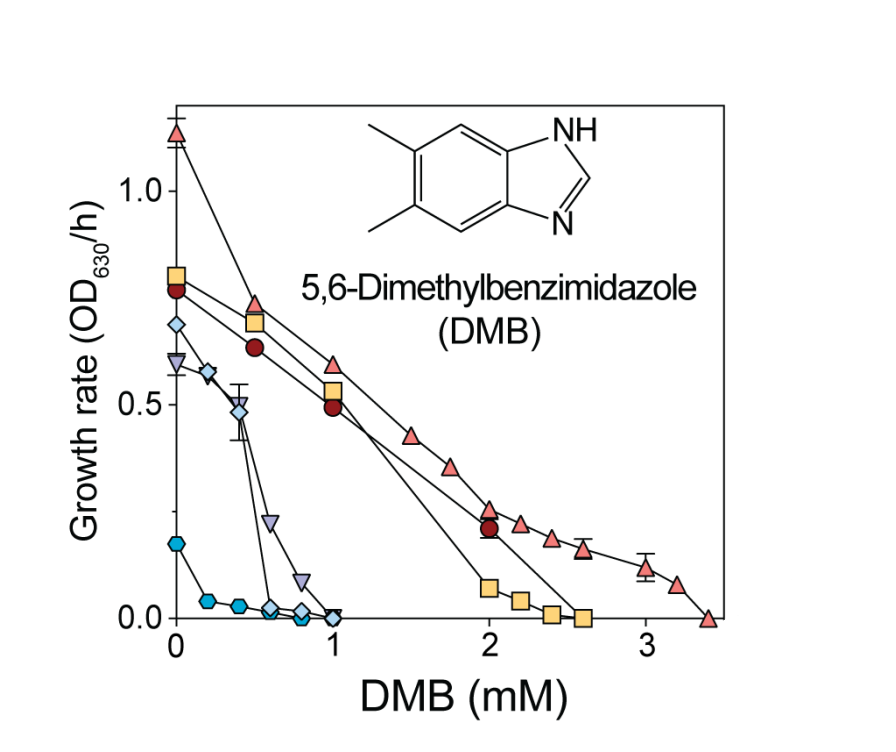
The coenzyme B12 precursor 5,6-dimethylbenzimidazole is a flavin antagonist in Salmonella
Lahiru Malalasekara1 and Jorge C. Escalante-Semerena1,*
Here we investigated why 5,6-dimethylbenzimidazole (DMB) inhibits in S. Typhimurium. Briefly, we determined that the structural similarities of the substituted benzene ring of DMB with the isoalloxazine moiety of flavins is responsible for the deleterious effects of this CoB12 precursor.
Increased Trypanosoma brucei cathepsin-L activity inhibits human serum-mediated trypanolysis
Sam Alsford
This article comments on work published by Alsford et al. (PLoS Pathogens, 2014), which identified a Trypanosoma brucei lysosomal cathepsin with an inhibitory effect on human serum’s trypanolytic action.
A novel role of centrin in flagellar motility: stabilizing an inner-arm dynein motor in the flagellar axoneme
Ziyin Li
This article comments on work published by Wei et al. (Nat Comm, 2014), which discovered that centrin maintains the stability of an inner-arm dynein in the flagellar axoneme in Trypanosoma brucei.
A non-proteolytic function of ubiquitin in transcription repression
Ada Ndoja and Tingting Yao
This article comments on work published by Ndoja et al. (Mol Cell, 2014), which demonstrates that monoubiquitination of some transcription activators can inhibit transcription by recruiting the AAA+ ATPase Cdc48 (also known in metazoans as p97 or VCP), which then extracts the ubiquitinated activator from DNA.
Mutagenesis by host antimicrobial peptides: insights into microbial evolution during chronic infections
Dominique H. Limoli and Daniel J. Wozniak
This article comments on work published by Limoli et al. ((PLoS Pathogens, 2014), which provides evidence that at subinhibitory levels, AMPs promote mutations in bacterial DNA, which enhance bacterial survival.
Where antibiotic resistance mutations meet quorum-sensing
Rok Krašovec1, Roman V. Belavkin2, John A.D. Aston3, Alastair Channon4, Elizabeth Aston4, Bharat M. Rash1, Manikandan Kadirvel5,6, Sarah Forbes6, and Christopher G. Knight1
This article comments on work published by Krašovec et al. (Nat Comm, 2014), which found that the modulation of de novo mutation to promote antibiotic resistance depends on the density of the bacterial population and cell-cell interactions (rather than, for instance, the level of stress).
Sphingolipids and mitochondrial function, lessons learned from yeast
Pieter Spincemaille1, Bruno P.A. Cammue1,2 and Karin Thevissen1
This article reviews recent research showing that Saccharomyces cerevisiae is an invaluable model to investigate sphingolipids as signaling molecules in modulating mitochondrial function, but can also be used as a tool to further enhance our current knowledge on sphingolipids and mitochondria in mammalian cells.
Genome evolution in yeast reveals connections between rare mutations in human cancer
Xinchen Teng1,2 and J. Marie Hardwick2
This article comments on work published by Teng et al. (Mol Cell, 2013), which, using the yeast knockout collections, provides hard evidence that single gene deletions/mutations in most non-essential genes can drive the selection for cancer-like mutations.
Decoding the biosynthesis and function of diphthamide, an enigmatic modification of translation elongation factor 2 (EF2)
Raffael Schaffrath and Michael J. R. Stark
This article comments on work published by Uthman et al. (PLoS Genet, 2013), which suggests that Dph5 has a novel role as an EF2 inhibitor that affects cell growth when diphthamide synthesis is blocked or incomplete and shows that diphthamide promotes the accuracy of EF2 performance during translation.
The emerging role of complex modifications of tRNALysUUU in signaling pathways
Patrick C. Thiaville1,2,3,4 and Valérie de Crécy-Lagard2,4
This comment discusses the article “Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling” by Scheidt et al (Microbial Cell, 2014).
Only functional localization is faithful localization
Roland Lill1,2,3
This article comments on work published by Peleh et al. (Microbial Cell 2014), which analyzes the localization of Dre2 in Saccharomyces cerevisiae.
One cell, one love: a journal for microbial research
Didac Carmona-Gutierrez1, Guido Kroemer2-6 and Frank Madeo1
In this inaugural article of Microbial Cell, we highlight the importance of microbial research in general and the journal’s intention to serve as a publishing forum that supports and enfolds the scientific diversity in this area as it provides a unique, high-quality and universally accessible source of information and inspiration.
What’s the role of autophagy in trypanosomes?
Katherine Figarella1 and Néstor L. Uzcátegui1,2
This article comments on Proto et al. (Microbial Cell, 2014), who report first insights into the molecular mechanism of autophagy in African trypanosomes by generating reporter bloodstream form cell lines.
Microbial Cell
is an open-access, peer-reviewed journal that publishes exceptionally relevant research works that implement the use of unicellular organisms (and multicellular microorganisms) to understand cellular responses to internal and external stimuli and/or human diseases.
you can trust
Can’t find what you’re looking for?
You can browse all our issues and published articles here.
FAQs
Peer-reviewed, open-access research using unicellular organisms (and multicellular microorganisms) to understand cellular responses and human disease.
The journal (founded in 2014) is led by its Editors-in-Chief Frank Madeo, Didac Carmona-Gutierrez, and Guido Kroemer
Microbial Cell has been publishing original scientific literature since 2014, and from the very beginning has been managed by active scientists through an independent Publishing House (Shared science Publishers). The journal was conceived as a platform to acknowledge the importance of unicellular organisms, both as model systems as well as in the biological context of human health and disease.
Ever since, Microbial Cell has very positively developed and strongly grown into a respected journal in the unicellular research community and even beyond. This scientific impact is reflected in the yearly number of citations obtained by articles published in Microbial Cell, as recorded by the Web of Science (Clarivate, formerly Thomson/Reuters):

The scientific impact of Microbial Cell is also mirrored in a series of milestones:
2015: Microbial Cell is included in the Emerging Sources Citation Index (ESCI), a selection of developing journals drafted by Clarivate Analytics based on the candidate’s publishing standards, quality, editorial content, and citation data. Note: As an ESCI-selected journal, Microbial Cell is currently being evaluated in a rigorous and long process to determine an inclusion in the Science Citation Index Expanded (SCIE), which allows the official calculation of Clarivate Analytics’ impact factor.
2016: Microbial Cell is awarded the so-called DOAJ Seal by the selective Directory of Open Access Journals (DOAJ). The DOAJ Seal is an exclusive mark of certification for open access journals granted by DOAJ to journals that adhere to outstanding best practice and achieve an extra high and clear commitment to open access and high publishing standards.
2017: Microbial Cell is included in Pubmed Central (PMC), allowing the archiving of all the journal’s articles in PMC and PubMed.
2019: Microbial Cell is indexed in the prestigious abstract and citation database Scopus after a thorough selection process. This also means that Microbial Cell obtains, for the first time, an official Scopus CiteScore as well as an official journal ranking in the Scimago Journal and Country Ranking.
2022: Microbial Cell’s CiteScore reaches a value of 7.2 for the year 2021, positioning Microbial Cell among the top microbiology journals (previously available CiteScores: 2019: 5.4; 2020: 5.1).
2022: Microbial Cell is indexed in the highly selective Science Citation Index Expanded™, which covers approx. 9,500 of the world’s most impactful journals across 178 scientific disciplines. In their journal selection and curation process, Clarivate´s editors apply 24 ‘quality’ criteria and four ‘impact’ criteria to select the most influential journals in their respective fields. This selection is also a pre-requisite for inclusion in the JCR, which features the impact factor.
2022: Microbial Cell is listed in the Journal Citation Reports™ (JCR), and obtains its first official Journal Impact Factor™ (JIF) for the year 2021: 5.316.Check Article Types and Manuscript Preparation guidelines. Submit online via Scholastica.





Metabolic pathways further increase the complexity of cell size control in budding yeast
Jorrit M. Enserink
This article comments on work published by Soma et al. (Microbial Cell, 2014), which teased apart the effect of metabolism and growth rate on setting of critical cell size in Saccharomyces cerevisiae.