
Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.

Luminal acetylation of microtubules is not essential for Plasmodium berghei and Toxoplasma gondii survival
Acetylation of α-tubulin at lysine 40 is not essential for cytoskeletal stability in Plasmodium berghei or Toxoplasma gondii, suggesting redundancy and plasticity in microtubule regulation in these parasites.

The dual-site agonist for human M2 muscarinic receptors Iper-8-naphtalimide induces mitochondrial dysfunction in Saccharomyces cerevisiae
S. cerevisiae is a model to study human GPCRs. N-8-Iper, active against glioblastoma via M2 receptor, causes mitochondrial damage in yeast by binding Ste2, highlighting evolutionary conservation of GPCRs.

Integrative Omics reveals changes in the cellular landscape of peroxisome-deficient pex3 yeast cells
To uncover the consequences of peroxisome deficiency, we compared Saccharomyces cerevisiae wild-type with pex3 cells, which lack peroxisomes, employing quantitative proteomics and transcriptomics technologies.

Regulation of extracellular vesicles for protein secretion in Aspergillus nidulans
Rebekkah E. Pope1, Patrick Ballmann2, Lisa Whitworth3 and Rolf A. Prade1,*
This study reveals that Aspergillus nidulans boosts extracellular vesicle production when ER-trafficked enzymes are induced, uncovering how fungi remodel their secretome through vesicle-mediated secretion to adapt to changing environments and biofilm formation.

Transcriptomic response to different heme sources in Trypanosoma cruzi epimastigotes
Evelyn Tevere1,a, María G. Mediavilla1,a, Cecilia B. Di Capua1, Marcelo L. Merli1, Carlos Robello2,3, Luisa Berná2,4 and Julia A. Cricco
This study uncovers how the Chagas disease parasite adapts to changes in heme, an essential molecule for its survival, providing transcriptional clues to heme metabolism and identifying a previously unreported heme-binding protein in T. cruzi.
Sir2 regulates selective autophagy in stationary-phase yeast cells
Ji-In Ryua, Juhye Junga, and Jeong-Yoon Kim
This study establishes Sir2 as a previously unrecognized regulator of selective autophagy during the stationary phase and highlight how cells dynamically control organelle degradation.
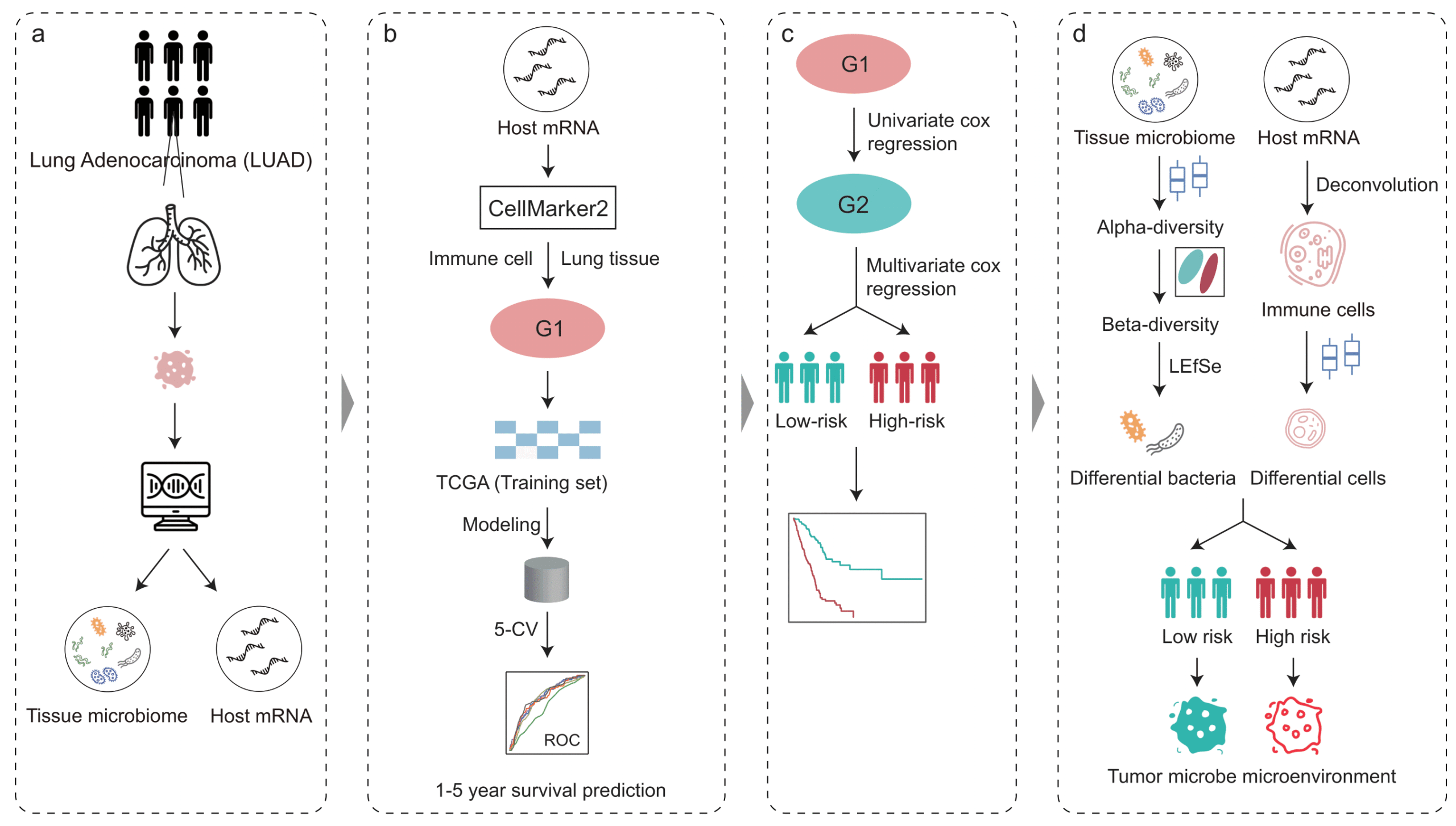
Predictable regulation of survival by intratumoral microbe-immune crosstalk in patients with lung adenocarcinoma
Shuo Shi1, Yuwen Chu2,3, Haiyan Liu4,5, Lan Yu6,7,8, Dejun Sun8,9, Jialiang Yang2,3,5, Geng Tian2,3, Lei Ji2,3, Cong Zhang10 and Xinxin Lu11
Intratumoral microbiota can regulate the tumor immune microenvironment (TIME) and mediate tumor prognosis by promoting inflammatory response or inhibiting anti-tumor effects. Our study demonstrated that intratumoral microbiota-immune crosstalk was strongly associated with prognosis in LUAD patients, which would provide new targets for the development of precise therapeutic strategies.

The last two transmembrane helices in the APC-type FurE transporter act as an intramolecular chaperone essential for concentrative ER-exit
Yiannis Pyrris1, Georgia F. Papadaki1, Emmanuel Mikros2 and George Diallinas1,3
FurE is a H+ symporter specific for the cellular uptake of uric acid, allantoin, uracil, and toxic nucleobase analogues in the fungus Aspergillus nidulans. Being member of the NCS1 protein family, FurE is structurally related to the APC-superfamily of transporters.

Basal level of ppGpp coordinates Escherichia coli cell heterogeneity and ampicillin resistance and persistence
Paulina Katarzyna Grucela1 and Yong Everett Zhang1
The universal stringent response alarmone ppGpp (guanosine penta and tetra phosphates) plays a crucial role in various aspects of fundamental cell physiology (e.g., cell growth rate, cell size) and thus bacterial tolerance to and survival of external stresses, including antibiotics. In tihs study, we discuss the fundamental role of basal level of ppGpp in regulating cell homogeneity and ampicillin persistence.
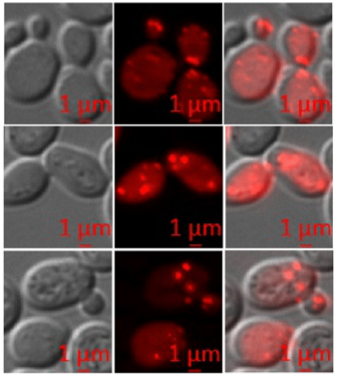
Investigation of the acetic acid stress response in Saccharomyces cerevisiae with mutated H3 residues
Nitu Saha1, Swati Swagatika1 and Raghuvir Singh Tomar1
Yeast cells respond to acetic acid in diverse ways. Here, we have elucidated the deleterious effects of acetic acid on different histone mutants
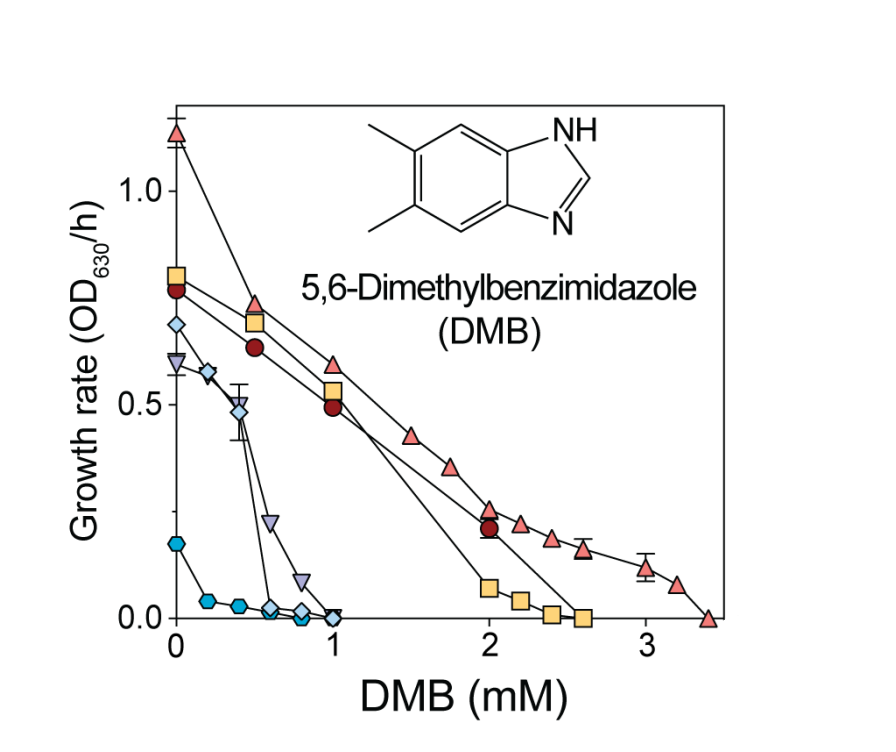
The coenzyme B12 precursor 5,6-dimethylbenzimidazole is a flavin antagonist in Salmonella
Lahiru Malalasekara1 and Jorge C. Escalante-Semerena1,*
Here we investigated why 5,6-dimethylbenzimidazole (DMB) inhibits in S. Typhimurium. Briefly, we determined that the structural similarities of the substituted benzene ring of DMB with the isoalloxazine moiety of flavins is responsible for the deleterious effects of this CoB12 precursor.
A novel mechanism involved in the coupling of mitochondrial biogenesis to oxidative phosphorylation
Jelena Ostojić1, Jean-Paul di Rago2,3, Geneviève Dujardin1,*
This article comments on a study by Ostojić et al. (Cell Metabolism, 2013), which has uncovered a regulatory loop by which the biogenesis of a major enzyme of the OXPHOS pathway, the respiratory complex III, is coupled to the energy producing activity of the mitochondria.
Identifying the assembly pathway of cyanophage inside the marine bacterium using electron cryo-tomography
Wei Dai1, Michael F. Schmid1, Jonathan A. King2, Wah Chiu1,*
Thiswork comments on a study by Dai et al. (Nature 2013) that illustrates that electron cryo-tomography is an approach whereby one can capture directly structural snapshots of transient phage assembly intermediates during maturation process. Such analysis can be generalizable not only to human viruses in human cells but also various molecular machines undergoing biological processes.
Targeting GATA transcription factors – a novel strategy for anti-aging interventions?
Andreas Zimmermann1, Katharina Kainz1,2, Sebastian J. Hofer1,3, Maria A. Bauer1, Sabrina Schroeder1, Jörn Dengjel4, Federico Pietrocola5, Oliver Kepp6-9, Christoph Ruckenstuhl1, Tobias Eisenberg1,3,10,11, Stephan J. Sigrist12, Frank Madeo1,3,10, Guido Kroemer6-9, 13-15 and Didac Carmona-Gutierrez1
This article comments on work published by Carmona-Gutierrez et al. (Nat Commun., 2019), which identified a natural compound, 4,4′-dimethoxychalcone, inducing autophagy and prolonging lifespan in different organisms through a mechanism that involves GATA transcription factors.
In the beginning was the word: How terminology drives our understanding of endosymbiotic organelles
Miroslav Oborník 1,2
This In the Pit article argues that the naming conventions for biological entities influence research perspectives and methodologies, advocating for mitochondria and plastids to be classified and named as bacteria due to their endosymbiotic origins, with potential implications for our understanding of bacterial prevalence, definitions of the microbiome and multicellularity, and the concept of endosymbiotic domestication.
What’s in a name? How organelles of endosymbiotic origin can be distinguished from endosymbionts
Ansgar Gruber1
This In the Pit article suggests redefining the relationship between hosts and endosymbionts, like mitochondria and plastids, as a single species based on “sexual symbiont integration,” the loss of independent speciation, and congruence in genetic recombination and population sizes, rather than solely on historic classifications or structural properties.
Microbial wars: competition in ecological niches and within the microbiome
Maria A. Bauer1, Katharina Kainz1, Didac Carmona-Gutierrez1 and Frank Madeo1,2
In this Editorial Bauer et al. provide a brief overview on microbial competition and discuss some of its roles and consequences that directly affect humans.
Exploring the mechanism of amebic trogocytosis: the role of amebic lysosomes
Allissia A. Gilmartin1 and William A. Petri, Jr1,2,3
In this article, the authors comment on the study “Inhibition of Amebic Lysosomal Acidification Blocks Amebic Trogocytosis and Cell Killing” by Gilmartin et al. (MBio, 2017), discussing the the role of amebic lysosomes in Trogocytosis, the intracellular transfer of fragments of cell material.
Uncovering the hidden: complexity and strategies for diagnosing latent tuberculosis
Mario Alberto Flores-Valdez
This editorial postulates that advanced proteomic and transcriptomic techniques are evolving and may enhance the detection of latent tuberculosis, thereby distinguishing true M. tuberculosis infections from other conditions, which is vital for controlling potential reactivation and transmission.
The Yin & Yang of Mitochondrial Architecture – Interplay of MICOS and F1Fo-ATP synthase in cristae formation
Heike Rampelt1 and Martin van der Laan2
This Editorial posits that mitochondrial cristae architecture is shaped by the interplay of MICOS and ATP synthase, with a recent study illuminating their roles in cristae formation and maintenance.
When a ribosomal protein grows up – the ribosome assembly path of Rps3
Brigitte Pertschy
This article comments on two papers by Mitterer et al., which followed yeast protein Rps3, highlighting the sophisticated mechanisms for protein protection, nuclear transport, and integration into pre-ribosomal particles for final assembly with 40S subunits.
Microbial Cell
is an open-access, peer-reviewed journal that publishes exceptionally relevant research works that implement the use of unicellular organisms (and multicellular microorganisms) to understand cellular responses to internal and external stimuli and/or human diseases.
you can trust
Can’t find what you’re looking for?
You can browse all our issues and published articles here.
FAQs
Peer-reviewed, open-access research using unicellular organisms (and multicellular microorganisms) to understand cellular responses and human disease.
The journal (founded in 2014) is led by its Editors-in-Chief Frank Madeo, Didac Carmona-Gutierrez, and Guido Kroemer
Microbial Cell has been publishing original scientific literature since 2014, and from the very beginning has been managed by active scientists through an independent Publishing House (Shared science Publishers). The journal was conceived as a platform to acknowledge the importance of unicellular organisms, both as model systems as well as in the biological context of human health and disease.
Ever since, Microbial Cell has very positively developed and strongly grown into a respected journal in the unicellular research community and even beyond. This scientific impact is reflected in the yearly number of citations obtained by articles published in Microbial Cell, as recorded by the Web of Science (Clarivate, formerly Thomson/Reuters):

The scientific impact of Microbial Cell is also mirrored in a series of milestones:
2015: Microbial Cell is included in the Emerging Sources Citation Index (ESCI), a selection of developing journals drafted by Clarivate Analytics based on the candidate’s publishing standards, quality, editorial content, and citation data. Note: As an ESCI-selected journal, Microbial Cell is currently being evaluated in a rigorous and long process to determine an inclusion in the Science Citation Index Expanded (SCIE), which allows the official calculation of Clarivate Analytics’ impact factor.
2016: Microbial Cell is awarded the so-called DOAJ Seal by the selective Directory of Open Access Journals (DOAJ). The DOAJ Seal is an exclusive mark of certification for open access journals granted by DOAJ to journals that adhere to outstanding best practice and achieve an extra high and clear commitment to open access and high publishing standards.
2017: Microbial Cell is included in Pubmed Central (PMC), allowing the archiving of all the journal’s articles in PMC and PubMed.
2019: Microbial Cell is indexed in the prestigious abstract and citation database Scopus after a thorough selection process. This also means that Microbial Cell obtains, for the first time, an official Scopus CiteScore as well as an official journal ranking in the Scimago Journal and Country Ranking.
2022: Microbial Cell’s CiteScore reaches a value of 7.2 for the year 2021, positioning Microbial Cell among the top microbiology journals (previously available CiteScores: 2019: 5.4; 2020: 5.1).
2022: Microbial Cell is indexed in the highly selective Science Citation Index Expanded™, which covers approx. 9,500 of the world’s most impactful journals across 178 scientific disciplines. In their journal selection and curation process, Clarivate´s editors apply 24 ‘quality’ criteria and four ‘impact’ criteria to select the most influential journals in their respective fields. This selection is also a pre-requisite for inclusion in the JCR, which features the impact factor.
2022: Microbial Cell is listed in the Journal Citation Reports™ (JCR), and obtains its first official Journal Impact Factor™ (JIF) for the year 2021: 5.316.Check Article Types and Manuscript Preparation guidelines. Submit online via Scholastica.





Sulfur dioxide resistance in Saccharomyces cerevisiae: beyond SSU1
Estéfani García-Ríos1 and José Manuel Guillamón1
This article discusses the importance of understanding sulfite resistance in Saccharomyces cerevisiae due to its use in winemaking and the potential role of the transcription factor Com2. While the SSU1 gene and its activity have been correlated with sulfite tolerance, the work by Lage et al. (2019) indicates that Com2 might control a large percentage of the genes activated by SO2 and contribute to the yeast’s protective response, offering new insights into the molecular factors influencing this oenological trait.